The COVID-19 immune landscape is dynamically and reversibly correlated with disease severity
- PMID: 33529167
- PMCID: PMC7843226
- DOI: 10.1172/JCI143648
The COVID-19 immune landscape is dynamically and reversibly correlated with disease severity
Abstract
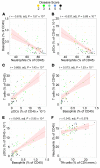
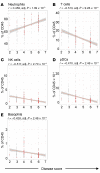
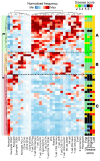
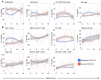
BACKGROUNDDespite a rapidly growing body of literature on coronavirus disease 2019 (COVID-19), our understanding of the immune correlates of disease severity, course, and outcome remains poor.METHODSUsing mass cytometry, we assessed the immune landscape in longitudinal whole-blood specimens from 59 patients presenting with acute COVID-19 and classified based on maximal disease severity. Hospitalized patients negative for SARS-CoV-2 were used as controls.RESULTSWe found that the immune landscape in COVID-19 formed 3 dominant clusters, which correlated with disease severity. Longitudinal analysis identified a pattern of productive innate and adaptive immune responses in individuals who had a moderate disease course, whereas those with severe disease had features suggestive of a protracted and dysregulated immune response. Further, we identified coordinate immune alterations accompanying clinical improvement and decline that were also seen in patients who received IL-6 pathway blockade.CONCLUSIONThe hospitalized COVID-19 negative cohort allowed us to identify immune alterations that were shared between severe COVID-19 and other critically ill patients. Collectively, our findings indicate that selection of immune interventions should be based in part on disease presentation and early disease trajectory due to the profound differences in the immune response in those with mild to moderate disease and those with the most severe disease.FUNDINGBenaroya Family Foundation, the Leonard and Norma Klorfine Foundation, Glenn and Mary Lynn Mounger, and the National Institutes of Health.
Keywords: Adaptive immunity; COVID-19; Innate immunity.
Conflict of interest statement
Figures






References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

