Pathogenic LMNA variants disrupt cardiac lamina-chromatin interactions and de-repress alternative fate genes
- PMID: 33529599
- PMCID: PMC8106635
- DOI: 10.1016/j.stem.2020.12.016
Pathogenic LMNA variants disrupt cardiac lamina-chromatin interactions and de-repress alternative fate genes
Abstract
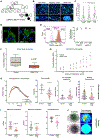
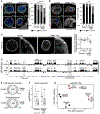
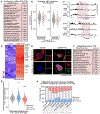
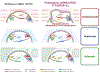
Pathogenic mutations in LAMIN A/C (LMNA) cause abnormal nuclear structure and laminopathies. These diseases have myriad tissue-specific phenotypes, including dilated cardiomyopathy (DCM), but how LMNA mutations result in tissue-restricted disease phenotypes remains unclear. We introduced LMNA mutations from individuals with DCM into human induced pluripotent stem cells (hiPSCs) and found that hiPSC-derived cardiomyocytes, in contrast to hepatocytes or adipocytes, exhibit aberrant nuclear morphology and specific disruptions in peripheral chromatin. Disrupted regions were enriched for transcriptionally active genes and regions with lower LAMIN B1 contact frequency. The lamina-chromatin interactions disrupted in mutant cardiomyocytes were enriched for genes associated with non-myocyte lineages and correlated with higher expression of those genes. Myocardium from individuals with LMNA variants similarly showed aberrant expression of non-myocyte pathways. We propose that the lamina network safeguards cellular identity and that pathogenic LMNA variants disrupt peripheral chromatin with specific epigenetic and molecular characteristics, causing misexpression of genes normally expressed in other cell types.
Keywords: genome organization; hiPSC; laminopathy; peripheral heterochromatin.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests K.M. is an advisor to and holds equity in Variant Bio and Verve Therapeutics.
Figures



References
-
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 57, 289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

