SARS-CoV-2 persistence is associated with antigen-specific CD8 T-cell responses
- PMID: 33530000
- PMCID: PMC7847186
- DOI: 10.1016/j.ebiom.2021.103230
SARS-CoV-2 persistence is associated with antigen-specific CD8 T-cell responses
Abstract
Background: Upon SARS-CoV-2 infection, most individuals develop neutralizing antibodies and T-cell immunity. However, some individuals reportedly remain SARS-CoV-2 PCR positive by pharyngeal swabs weeks after recovery. Whether viral RNA in these persistent carriers is contagious and stimulates SARS-CoV-2-specific immune responses is unknown.
Methods: This cohort study was conducted between April 3rd-July 9th 2020, recruiting COVID-19 recovered individuals that were symptom-free for at least 14 days. We collected serum for SARS-CoV-2-specific total Ig, IgA and IgM detection by ELISA, pharyngeal swabs (two time points) for ddPCR and PBMCs for anti-SARS-CoV-2 CD8 T-cell dextramer analyses.
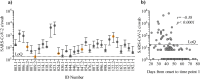
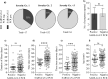
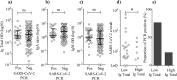
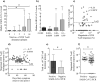
Findings: We enrolled 203 post-symptomatic participants with a previous RT-PCR-verified SARS-CoV-2 infection. At time point 1, a median of 23 days (range 15-44) after recovery, 26 individuals (12⋅8%) were PCR positive. At time point 2, 90 days (median, range 85-105) after recovery, 5 (5⋅3%) were positive. There was no difference in SARS-CoV-2 antibody levels between the PCR negative and positive group. The persistent PCR positive group however, had SARS-CoV-2-specific CD8 T-cell responses of significantly increased breadth and magnitude. Assisted contact tracing among persistent PCR positive individuals revealed zero new COVID-19 diagnoses among 757 close contacts.
Interpretation: Persistent pharyngeal SARS-CoV-2 PCR positivity in post-symptomatic individuals is associated with elevated cellular immune responses and thus, the viral RNA may represent replicating virus. However, transmission to close contacts was not observed indicating that persistent PCR positive individuals are not contagious at the post-symptomatic stage of the infection.
Keywords: Antibodies; CD8 T cell responses; Contact tracing; Immunology; Persistent PCR positive; SARS-CoV-2; Transmission.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors report no conflict of interest.
Figures
Comment in
-
Persistent viral RNA shedding in COVID-19: Caution, not fear.EBioMedicine. 2021 Feb;64:103234. doi: 10.1016/j.ebiom.2021.103234. Epub 2021 Feb 10. EBioMedicine. 2021. PMID: 33581642 Free PMC article. No abstract available.
References
-
- World Health Organization. COVID-19 weekly epidemiological update. World Heal Organ. 2020 December 27.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

