HIV-1: To Splice or Not to Splice, That Is the Question
- PMID: 33530363
- PMCID: PMC7912102
- DOI: 10.3390/v13020181
HIV-1: To Splice or Not to Splice, That Is the Question
Abstract
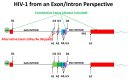
The transcription of the HIV-1 provirus results in only one type of transcript-full length genomic RNA. To make the mRNA transcripts for the accessory proteins Tat and Rev, the genomic RNA must completely splice. The mRNA transcripts for Vif, Vpr, and Env must undergo splicing but not completely. Genomic RNA (which also functions as mRNA for the Gag and Gag/Pro/Pol precursor polyproteins) must not splice at all. HIV-1 can tolerate a surprising range in the relative abundance of individual transcript types, and a surprising amount of aberrant and even odd splicing; however, it must not over-splice, which results in the loss of full-length genomic RNA and has a dramatic fitness cost. Cells typically do not tolerate unspliced/incompletely spliced transcripts, so HIV-1 must circumvent this cell policing mechanism to allow some splicing while suppressing most. Splicing is controlled by RNA secondary structure, cis-acting regulatory sequences which bind splicing factors, and the viral protein Rev. There is still much work to be done to clarify the combinatorial effects of these splicing regulators. These control mechanisms represent attractive targets to induce over-splicing as an antiviral strategy. Finally, splicing has been implicated in latency, but to date there is little supporting evidence for such a mechanism. In this review we apply what is known of cellular splicing to understand splicing in HIV-1, and present data from our newer and more sensitive deep sequencing assays quantifying the different HIV-1 transcript types.
Keywords: HIV-1; HIV-1 latency; HIV-1 oversplicing; HIV-1 splicing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Emery A. (Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC, USA), Archin N. (UNC HIV Cure Center, University of North Carolina, Chapel Hill, NC, USA), Margolis D. (UNC HIV Cure Center, University of North Carolina, Chapel Hill, NC, USA), Williamson C. (Division of Medical Virology, Institute of Infectious Disease and Molecular Medicine, University of Cape Town, Cape Town, South Africa), Garrett N. (Centre for the AIDS Programme of Research in South Africa, University of KwaZulu- Natal, Durban, South Africa). Splicing in HIV-1 outgrowth viruses from ART suppressed individuals. 2019.
-
- Ocwieja K.E., Sherrill-Mix S., Mukherjee R., Custers-Allen R., David P., Brown M., Wang S., Link D.R., Olson J., Travers K., et al. Dynamic regulation of HIV-1 mRNA populations analyzed by single-molecule enrichment and long-read sequencing. Nucleic Acids Res. 2012;40:10345–10355. doi: 10.1093/nar/gks753. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

