Decorating the surface of Escherichia coli with bacterial lipoproteins: a comparative analysis of different display systems
- PMID: 33531008
- PMCID: PMC7853708
- DOI: 10.1186/s12934-021-01528-z
Decorating the surface of Escherichia coli with bacterial lipoproteins: a comparative analysis of different display systems
Abstract
Background: The display of recombinant proteins on cell surfaces has a plethora of applications including vaccine development, screening of peptide libraries, whole-cell biocatalysts and biosensor development for diagnostic, industrial or environmental purposes. In the last decades, a wide variety of surface display systems have been developed for the exposure of recombinant proteins on the surface of Escherichia coli, such as autotransporters and outer membrane proteins.
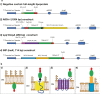
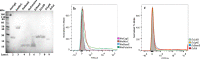
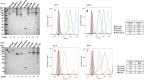
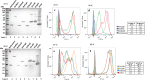
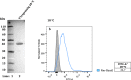
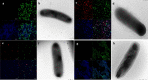
Results: In this study, we assess three approaches for the surface display of a panel of heterologous and homologous mature lipoproteins in E. coli: four from Neisseria meningitidis and four from the host strain that are known to be localised in the inner leaflet of the outer membrane. Constructs were made carrying the sequences coding for eight mature lipoproteins, each fused to the delivery portion of three different systems: the autotransporter adhesin involved in diffuse adherence-I (AIDA-I) from enteropathogenic E. coli, the Lpp'OmpA chimaera and a truncated form of the ice nucleation protein (INP), InaK-NC (N-terminal domain fused with C-terminal one) from Pseudomonas syringae. In contrast to what was observed for the INP constructs, when fused to the AIDA-I or Lpp'OmpA, most of the mature lipoproteins were displayed on the bacterial surface both at 37 and 25 °C as demonstrated by FACS analysis, confocal and transmission electron microscopy.
Conclusions: To our knowledge this is the first study that compares surface display systems using a number of passenger proteins. We have shown that the experimental conditions, including the choice of the carrier protein and the growth temperature, play an important role in the translocation of mature lipoproteins onto the bacterial surface. Despite all the optimization steps performed with the InaK-NC anchor motif, surface exposure of the passenger proteins used in this study was not achieved. For our experimental conditions, Lpp'OmpA chimaera has proved to be an efficient surface display system for the homologous passenger proteins although cell lysis and phenotype heterogeneity were observed. Finally, AIDA-I was found to be the best surface display system for mature lipoproteins (especially heterologous ones) in the E. coli host strain with no inhibition of growth and only limited phenotype heterogeneity.
Keywords: AIDA-I; Escherichia coli; Ice nucleation protein (InaK-NC); Lipoproteins; Lpp’OmpA chimaera; Surface display systems.
Conflict of interest statement
This work was sponsored by GlaxoSmithKline Biologicals SA. All authors have declared the following interests: MG, FG, LP, ID, DM and CB are employees of the GSK group of companies. CLG is a consultant for GSK, Italy. SN is a PhD student (University of Bologna) at GSK, Italy.
Figures
References
-
- Parola C, Mason DM, Zingg A, Neumeier D, Reddy ST. Genome engineering of hybridomas to generate stable cell lines for antibody expression. Methods Mol. Biol. 2018 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

