DNA methylation impact on Fabry disease
- PMID: 33531072
- PMCID: PMC7852133
- DOI: 10.1186/s13148-021-01019-3
DNA methylation impact on Fabry disease
Abstract
Background: Fabry disease (FD) is a rare X-linked disease caused by mutations in GLA gene with consequent lysosomal accumulation of globotriaosylceramide (Gb3). Women with FD often show highly heterogeneous symptoms that can manifest from mild to severe phenotype.
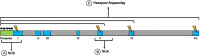
Main body: The phenotypic variability of the clinical manifestations in heterozygous women with FD mainly depends on the degree and direction of inactivation of the X chromosome. Classical approaches to measure XCI skewness might be not sufficient to explain disease manifestation in women. In addition to unbalanced XCI, allele-specific DNA methylation at promoter of GLA gene may influence the expression levels of the mutated allele, thus impacting the onset and the outcome of FD. In this regard, analyses of DNA methylation at GLA promoter, performed by approaches allowing distinction between mutated and non-mutated allele, may be much more informative. The aim of this review is to critically evaluate recent literature articles addressing the potential role of DNA methylation in the context of FD. Although up to date relatively few works have addressed this point, reviewing all pertinent studies may help to evaluate the importance of DNA methylation analysis in FD and to develop new research and technologies aimed to predict whether the carrier females will develop symptoms.
Conclusions: Relatively few studies have addressed the complexity of DNA methylation landscape in FD that remains poorly investigated. The hope for the future is that ad hoc and ultradeep methylation analyses of GLA gene will provide epigenetic signatures able to predict whether pre-symptomatic female carriers will develop symptoms thus helping timely interventions.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Desnick RJ, Wasserstein MP, Banikazemi M. Fabry disease (alpha-galactosidase A deficiency): renal involvement and enzyme replacement therapy. Contrib Nephrol. 2001;136:174–192. - PubMed
-
- Schiffmann R. Fabry disease. Handb Clin Neurol. 2015;132:231–248. - PubMed
-
- Meikle PJ, Hopwood JJ, Clague AE, Carey WF. Prevalence of lysosomal storage disorders. JAMA. 1999;281(3):249–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

