Integrative bioinformatics approaches for identifying potential biomarkers and pathways involved in non-obstructive azoospermia
- PMID: 33532314
- PMCID: PMC7844508
- DOI: 10.21037/tau-20-1029
Integrative bioinformatics approaches for identifying potential biomarkers and pathways involved in non-obstructive azoospermia
Abstract
Background: Non-obstructive azoospermia (NOA) is a disease related to spermatogenic disorders. Currently, the specific etiological mechanism of NOA is unclear. This study aimed to use integrated bioinformatics to screen biomarkers and pathways involved in NOA and reveal their potential molecular mechanisms.
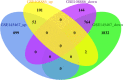
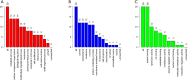
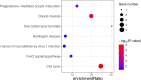
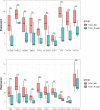
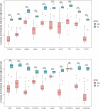
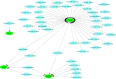
Methods: GSE145467 and GSE108886 gene expression profiles were obtained from the Gene Expression Omnibus (GEO) database. The differentially expressed genes (DEGs) between NOA tissues and matched obstructive azoospermia (OA) tissues were identified using the GEO2R tool. Common DEGs in the two datasets were screened out by the VennDiagram package. For the functional annotation of common DEGs, DAVID v.6.8 was used to perform Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. In accordance with data collected from the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database, a protein-protein interaction (PPI) network was constructed by Cytoscape. Cytohubba in Cytoscape was used to screen the hub genes. Furthermore, the hub genes were validated based on a separate dataset, GSE9210. Finally, potential micro RNAs (miRNAs) of hub genes were predicted by miRWalk 3.0.
Results: A total of 816 common DEGs, including 52 common upregulated and 764 common downregulated genes in two datasets, were screened out. Some of the more important of these pathways, including focal adhesion, PI3K-Akt signaling pathway, cell cycle, oocyte meiosis, AMP-activated protein kinase (AMPK) signaling pathway, FoxO signaling pathway, and Huntington disease, were involved in spermatogenesis. We further identified the top 20 hub genes from the PPI network, including CCNB2, DYNLL2, HMMR, NEK2, KIF15, DLGAP5, NUF2, TTK, PLK4, PTTG1, PBK, CEP55, CDKN3, CDC25C, MCM4, DNAI1, TYMS, PPP2R1B, DNAI2, and DYNLRB2, which were all downregulated genes. In addition, potential miRNAs of hub genes, including hsa-miR-3666, hsa-miR-130b-3p, hsa-miR-15b-5p, hsa-miR-6838-5p, and hsa-miR-195-5p, were screened out.
Conclusions: Taken together, the identification of the above hub genes, miRNAs and pathways will help us better understand the mechanisms associated with NOA, and provide potential biomarkers and therapeutic targets for NOA.
Keywords: Non-obstructive azoospermia (NOA); biomarkers; expression profiling data; functional enrichment analysis; protein–protein interactions.
2021 Translational Andrology and Urology. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1029). The authors have no conflicts of interest to declare.
Figures

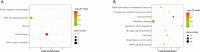
Similar articles
-
Integrative analyses of potential biomarkers and pathways for non-obstructive azoospermia.Front Genet. 2022 Nov 24;13:988047. doi: 10.3389/fgene.2022.988047. eCollection 2022. Front Genet. 2022. PMID: 36506310 Free PMC article.
-
Identification and Potential Value of Candidate Genes in Patients With Non-obstructive Azoospermia.Urology. 2022 Jun;164:133-139. doi: 10.1016/j.urology.2022.02.009. Epub 2022 Feb 24. Urology. 2022. PMID: 35219767
-
AKT3 and related molecules as potential biomarkers responsible for cryptorchidism and cryptorchidism-induced azoospermia.Transl Pediatr. 2021 Jul;10(7):1805-1817. doi: 10.21037/tp-21-31. Transl Pediatr. 2021. PMID: 34430428 Free PMC article.
-
Common gene signatures and key pathways in hypopharyngeal and esophageal squamous cell carcinoma: Evidence from bioinformatic analysis.Medicine (Baltimore). 2020 Oct 16;99(42):e22434. doi: 10.1097/MD.0000000000022434. Medicine (Baltimore). 2020. PMID: 33080677 Free PMC article.
-
STAT4 and COL1A2 are potential diagnostic biomarkers and therapeutic targets for heart failure comorbided with depression.Brain Res Bull. 2022 Jun 15;184:68-75. doi: 10.1016/j.brainresbull.2022.03.014. Epub 2022 Mar 31. Brain Res Bull. 2022. PMID: 35367598 Review.
Cited by
-
Constructing a seventeen-gene signature model for non-obstructive azoospermia based on integrated transcriptome analyses and WGCNA.Reprod Biol Endocrinol. 2023 Mar 21;21(1):30. doi: 10.1186/s12958-023-01079-5. Reprod Biol Endocrinol. 2023. PMID: 36945018 Free PMC article.
-
Genome-Wide Association Analyses in Family Triads and Dyads Following Assisted Reproductive Technology.Genet Epidemiol. 2025 Jul;49(5):e70011. doi: 10.1002/gepi.70011. Genet Epidemiol. 2025. PMID: 40457613 Free PMC article.
-
MicroRNA-targeting in male infertility: Sperm microRNA-19a/b-3p and its spermatogenesis related transcripts content in men with oligoasthenozoospermia.Front Cell Dev Biol. 2022 Sep 21;10:973849. doi: 10.3389/fcell.2022.973849. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36211460 Free PMC article.
-
Common as well as unique methylation-sensitive DNA regulatory elements in three mammalian SLC9C1 genes.Gene. 2024 Jan 30;893:147897. doi: 10.1016/j.gene.2023.147897. Epub 2023 Oct 11. Gene. 2024. PMID: 37832806 Free PMC article.
-
A Comparative Cross-Platform Analysis to Identify Potential Biomarker Genes for Evaluation of Teratozoospermia and Azoospermia.Genes (Basel). 2022 Sep 25;13(10):1721. doi: 10.3390/genes13101721. Genes (Basel). 2022. PMID: 36292606 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
