Prediction of Late-Onset Sepsis in Preterm Infants Using Monitoring Signals and Machine Learning
- PMID: 33532727
- PMCID: PMC7846455
- DOI: 10.1097/CCE.0000000000000302
Prediction of Late-Onset Sepsis in Preterm Infants Using Monitoring Signals and Machine Learning
Abstract
Objectives: Prediction of late-onset sepsis (onset beyond day 3 of life) in preterm infants, based on multiple patient monitoring signals 24 hours before onset.
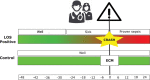
Design: Continuous high-resolution electrocardiogram and respiration (chest impedance) data from the monitoring signals were extracted and used to create time-interval features representing heart rate variability, respiration, and body motion. For each infant with a blood culture-proven late-onset sepsis, a Cultures, Resuscitation, and Antibiotics Started Here moment was defined. The Cultures, Resuscitation, and Antibiotics Started Here moment served as an anchor point for the prediction analysis. In the group with controls (C), an "equivalent crash moment" was calculated as anchor point, based on comparable gestational and postnatal age. Three common machine learning approaches (logistic regressor, naive Bayes, and nearest mean classifier) were used to binary classify samples of late-onset sepsis from C. For training and evaluation of the three classifiers, a leave-k-subjects-out cross-validation was used.
Setting: Level III neonatal ICU.
Patients: The patient population consisted of 32 premature infants with sepsis and 32 age-matched control patients.
Interventions: No interventions were performed.
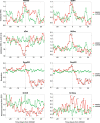
Measurements and main results: For the interval features representing heart rate variability, respiration, and body motion, differences between late-onset sepsis and C were visible up to 5 hours preceding the Cultures, Resuscitation, and Antibiotics Started Here moment. Using a combination of all features, classification of late-onset sepsis and C showed a mean accuracy of 0.79 ± 0.12 and mean precision rate of 0.82 ± 0.18 3 hours before the onset of sepsis.
Conclusions: Information from routine patient monitoring can be used to predict sepsis. Specifically, this study shows that a combination of electrocardiogram-based, respiration-based, and motion-based features enables the prediction of late-onset sepsis hours before the clinical crash moment.
Keywords: infant; intensive care units; machine learning; monitoring; neonatal; physiologic; predictive value of tests; premature; sepsis.
Copyright © 2021 The Authors. Published by Wolters Kluwer Health, Inc. on behalf of the Society of Critical Care Medicine.
Conflict of interest statement
The authors have disclosed that they do not have any potential conflicts of interest.
Figures

References
-
- Mahieu LM, Buitenweg N, Beutels P, et al. Additional hospital stay and charges due to hospital-acquired infections in a neonatal intensive care unit. J Hosp Infect. 2001; 47:223–229 - PubMed
-
- Stoll BJ, Hansen NI, Adams-Chapman I, et al. ; National Institute of Child Health and Human Development Neonatal Research Network. Neurodevelopmental and growth impairment among extremely low-birth-weight infants with neonatal infection. JAMA. 2004; 292:2357–2365 - PubMed
-
- Shane AL, Sánchez PJ, Stoll BJ. Neonatal sepsis. Lancet. 2017; 390:1770–1780 - PubMed
-
- Schelonka RL, Chai MK, Yoder BA, et al. Volume of blood required to detect common neonatal pathogens. J Pediatr. 1996; 129:275–278 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

