This is a preprint.
Synthetic nanobody-SARS-CoV-2 receptor-binding domain structures identify distinct epitopes
- PMID: 33532775
- PMCID: PMC7852268
- DOI: 10.1101/2021.01.27.428466
Synthetic nanobody-SARS-CoV-2 receptor-binding domain structures identify distinct epitopes
Update in
-
Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.J Biol Chem. 2021 Oct;297(4):101202. doi: 10.1016/j.jbc.2021.101202. Epub 2021 Sep 16. J Biol Chem. 2021. PMID: 34537245 Free PMC article.
Abstract
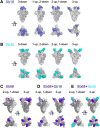
The worldwide spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) demands unprecedented attention. We report four X-ray crystal structures of three synthetic nanobodies (sybodies) (Sb16, Sb45 and Sb68) bind to the receptor-binding domain (RBD) of SARS-CoV-2: binary complexes of Sb16-RBD and Sb45-RBD; a ternary complex of Sb45-RBD-Sb68; and Sb16 unliganded. Sb16 and Sb45 bind the RBD at the ACE2 interface, positioning their CDR2 and CDR3 loops diametrically. Sb16 reveals a large CDR2 shift when binding the RBD. Sb68 interacts peripherally at the ACE2 interface; steric clashes with glycans explain its mechanism of viral neutralization. Superposing these structures onto trimeric spike (S) protein models indicates these sybodies bind conformations of the mature S protein differently, which may aid therapeutic design.
One sentence summary: X-ray structures of synthetic nanobodies complexed with the receptor-binding domain of the spike protein of SARS-CoV-2 reveal details of CDR loop interactions in recognition of distinct epitopic sites.
Conflict of interest statement
Figures



References
-
- Wibmer C. K. et al. , SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv, 2021.2001.2018.427166 (2021). - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
