This is a preprint.
Effect of mutations in the SARS-CoV-2 spike protein on protein stability, cleavage, and cell-cell fusion function
- PMID: 33532777
- PMCID: PMC7852270
- DOI: 10.1101/2021.01.24.428007
Effect of mutations in the SARS-CoV-2 spike protein on protein stability, cleavage, and cell-cell fusion function
Update in
-
Effect of clinical isolate or cleavage site mutations in the SARS-CoV-2 spike protein on protein stability, cleavage, and cell-cell fusion.J Biol Chem. 2021 Jul;297(1):100902. doi: 10.1016/j.jbc.2021.100902. Epub 2021 Jun 20. J Biol Chem. 2021. PMID: 34157282 Free PMC article.
Abstract
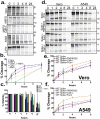
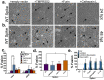
The SARS-CoV-2 spike protein (S) is the sole viral protein responsible for both viral binding to a host cell and the membrane fusion event needed for cell entry. In addition to facilitating fusion needed for viral entry, S can also drive cell-cell fusion, a pathogenic effect observed in the lungs of SARS-CoV-2 infected patients. While several studies have investigated S requirements involved in viral particle entry, examination of S stability and factors involved in S cell-cell fusion remain limited. We demonstrate that S must be processed at the S1/S2 border in order to mediate cell-cell fusion, and that mutations at potential cleavage sites within the S2 subunit alter S processing at the S1/S2 border, thus preventing cell-cell fusion. We also identify residues within the internal fusion peptide and the cytoplasmic tail that modulate S cell-cell fusion. Additionally, we examine S stability and protein cleavage kinetics in a variety of mammalian cell lines, including a bat cell line related to the likely reservoir species for SARS-CoV-2, and provide evidence that proteolytic processing alters the stability of the S trimer. This work therefore offers insight into S stability, proteolytic processing, and factors that mediate S cell-cell fusion, all of which help give a more comprehensive understanding of this highly sought-after therapeutic target.
Conflict of interest statement
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Figures


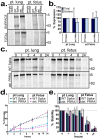
References
-
- World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. 2020. [cited 2020 12/2/2020]; Available from: https://covid19.who.int/.
-
- Lan J., et al. , Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature, 2020. 581(7807): p. 215–220. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
