The leucine-rich repeats in allelic barley MLA immune receptors define specificity towards sequence-unrelated powdery mildew avirulence effectors with a predicted common RNase-like fold
- PMID: 33534797
- PMCID: PMC7857584
- DOI: 10.1371/journal.ppat.1009223
The leucine-rich repeats in allelic barley MLA immune receptors define specificity towards sequence-unrelated powdery mildew avirulence effectors with a predicted common RNase-like fold
Abstract
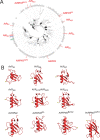
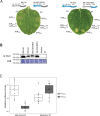
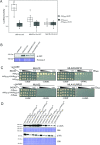
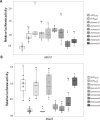
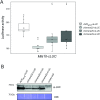
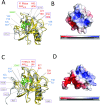
Nucleotide-binding domain leucine-rich repeat-containing receptors (NLRs) in plants can detect avirulence (AVR) effectors of pathogenic microbes. The Mildew locus a (Mla) NLR gene has been shown to confer resistance against diverse fungal pathogens in cereal crops. In barley, Mla has undergone allelic diversification in the host population and confers isolate-specific immunity against the powdery mildew-causing fungal pathogen Blumeria graminis forma specialis hordei (Bgh). We previously isolated the Bgh effectors AVRA1, AVRA7, AVRA9, AVRA13, and allelic AVRA10/AVRA22, which are recognized by matching MLA1, MLA7, MLA9, MLA13, MLA10 and MLA22, respectively. Here, we extend our knowledge of the Bgh effector repertoire by isolating the AVRA6 effector, which belongs to the family of catalytically inactive RNase-Like Proteins expressed in Haustoria (RALPHs). Using structural prediction, we also identified RNase-like folds in AVRA1, AVRA7, AVRA10/AVRA22, and AVRA13, suggesting that allelic MLA recognition specificities could detect structurally related avirulence effectors. To better understand the mechanism underlying the recognition of effectors by MLAs, we deployed chimeric MLA1 and MLA6, as well as chimeric MLA10 and MLA22 receptors in plant co-expression assays, which showed that the recognition specificity for AVRA1 and AVRA6 as well as allelic AVRA10 and AVRA22 is largely determined by the receptors' C-terminal leucine-rich repeats (LRRs). The design of avirulence effector hybrids allowed us to identify four specific AVRA10 and five specific AVRA22 aa residues that are necessary to confer MLA10- and MLA22-specific recognition, respectively. This suggests that the MLA LRR mediates isolate-specific recognition of structurally related AVRA effectors. Thus, functional diversification of multi-allelic MLA receptors may be driven by a common structural effector scaffold, which could be facilitated by proliferation of the RALPH effector family in the pathogen genome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Dyck PL, Kerber ER. Resistance of the Race-Specific Type. Cereal rusts. 1985;II.
-
- Flor HH. Inheritance of pathogenicity in Melampsora lini. Phytopathology; 1942. pp. 653–669.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

