Uncovering potential host proteins and pathways that may interact with eukaryotic short linear motifs in viral proteins of MERS, SARS and SARS2 coronaviruses that infect humans
- PMID: 33534852
- PMCID: PMC7857568
- DOI: 10.1371/journal.pone.0246150
Uncovering potential host proteins and pathways that may interact with eukaryotic short linear motifs in viral proteins of MERS, SARS and SARS2 coronaviruses that infect humans
Abstract
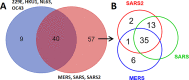
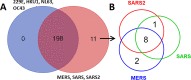
A coronavirus pandemic caused by a novel coronavirus (SARS-CoV-2) has spread rapidly worldwide since December 2019. Improved understanding and new strategies to cope with novel coronaviruses are urgently needed. Viruses (especially RNA viruses) encode a limited number and size (length of polypeptide chain) of viral proteins and must interact with the host cell components to control (hijack) the host cell machinery. To achieve this goal, the extensive mimicry of SLiMs in host proteins provides an effective strategy. However, little is known regarding SLiMs in coronavirus proteins and their potential targets in host cells. The objective of this study is to uncover SLiMs in coronavirus proteins that are present within host cells. These SLiMs have a high possibility of interacting with host intracellular proteins and hijacking the host cell machinery for virus replication and dissemination. In total, 1,479 SLiM hits were identified in the 16 proteins of 590 coronaviruses infecting humans. Overall, 106 host proteins were identified that may interact with SLiMs in 16 coronavirus proteins. These SLiM-interacting proteins are composed of many intracellular key regulators, such as receptors, transcription factors and kinases, and may have important contributions to virus replication, immune evasion and viral pathogenesis. A total of 209 pathways containing proteins that may interact with SLiMs in coronavirus proteins were identified. This study uncovers potential mechanisms by which coronaviruses hijack the host cell machinery. These results provide potential therapeutic targets for viral infections.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

