A comprehensive evaluation of skin aging-related circular RNA expression profiles
- PMID: 33534927
- PMCID: PMC8059755
- DOI: 10.1002/jcla.23714
A comprehensive evaluation of skin aging-related circular RNA expression profiles
Abstract
Background: Circular RNAs (circRNAs) have been shown to play important regulatory roles in a range of both pathological and physiological contexts, but their functions in the context of skin aging remain to be clarified. In the present study, we therefore, profiled circRNA expression profiles in four pairs of aged and non-aged skin samples to identify identifying differentially expressed circRNAs that may offer clinical value as biomarkers of the skin aging process.
Methods: We utilized an RNA-seq to profile the levels of circRNAs in eyelid tissue samples, with qRT-PCR being used to confirm these RNA-seq results, and with bioinformatics approaches being used to predict downstream target miRNAs for differentially expressed circRNAs.
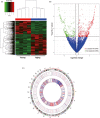
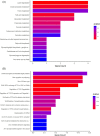
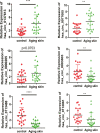
Results: In total, we identified 571 circRNAs with 348 and 223 circRNAs being up and downregulated that were differentially expressed in aged skin samples compared to young skin samples. The top 10 upregulated circRNAs in aged skin sample were hsa_circ_0123543, hsa_circ_0057742, hsa_circ_0088179, hsa_circ_0132428, hsa_circ_0094423, hsa_circ_0008166, hsa_circ_0138184, hsa_circ_0135743, hsa_circ_0114119, and hsa_circ_0131421. The top 10 reduced circRNAs were hsa_circ_0101479, hsa_circ_0003650, hsa_circ_0004249, hsa_circ_0030345, hsa_circ_0047367, hsa_circ_0055629, hsa_circ_0062955, hsa_circ_0005305, hsa_circ_0001627, and hsa_circ_0008531. Functional enrichment analyses revealed the potential functionality of these differentially expressed circRNAs. The top 3 enriched gene ontology (GO) terms of the host genes of differentially expressed circRNAs are regulation of GTPase activity, positive regulation of GTPase activity and autophagy. The top 3 enriched KEGG pathway ID are Lysine degradation, Fatty acid degradation and Inositol phosphate metabolism. The top 3 enriched reactome pathway ID are RAB GEFs exchange GTP for GDP on RABs, Regulation of TP53 Degradation and Regulation of TP53 Expression and Degradation. Six circRNAs were selected for qRT-PCR verification, of which 5 verification results were consistent with the sequencing results. Moreover, targeted miRNAs, such as hsa-miR-588, hsa-miR-612, hsa-miR-4487, hsa-miR-149-5p, hsa-miR-494-5p were predicted for circRna-miRna interaction networks.
Conclusion: Overall, these results offer new insights into circRNA expression profiles, potentially highlighting future avenues for research regarding the roles of these circRNAs in the context of skin aging.
Keywords: NGS; biomarker; circRNA; skin aging.
© 2021 The Authors. Journal of Clinical Laboratory Analysis published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare they have no competing interests.
Figures


References
-
- Lei L, Ou L, Yu X. The antioxidant effect of Asparagus cochinchinensis (Lour.) Merr. shoot in D‐galactose induced mice aging model and in vitro. J Chin Med Assoc. 2016;79(4):205‐211. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

