The In Silico Identification of Potential Members of the Ded1/DDX3 Subfamily of DEAD-Box RNA Helicases from the Protozoan Parasite Leishmania infantum and Their Analyses in Yeast
- PMID: 33535521
- PMCID: PMC7912733
- DOI: 10.3390/genes12020212
The In Silico Identification of Potential Members of the Ded1/DDX3 Subfamily of DEAD-Box RNA Helicases from the Protozoan Parasite Leishmania infantum and Their Analyses in Yeast
Abstract
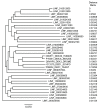
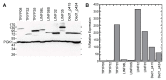
DEAD-box RNA helicases are ubiquitous proteins found in all kingdoms of life and that are associated with all processes involving RNA. Their central roles in biology make these proteins potential targets for therapeutic or prophylactic drugs. The Ded1/DDX3 subfamily of DEAD-box proteins is of particular interest because of their important role(s) in translation. In this paper, we identified and aligned the protein sequences of 28 different DEAD-box proteins from the kinetoplast-protozoan parasite Leishmania infantum, which is the cause of the visceral form of leishmaniasis that is often lethal if left untreated, and compared them with the consensus sequence derived from DEAD-box proteins in general, and from the Ded1/DDX3 subfamily in particular, from a wide variety of other organisms. We identified three potential homologs of the Ded1/DDX3 subfamily and the equivalent proteins from the related protozoan parasite Trypanosoma brucei, which is the causative agent of sleeping sickness. We subsequently tested these proteins for their ability to complement a yeast strain deleted for the essential DED1 gene. We found that the DEAD-box proteins from Trypanosomatids are highly divergent from other eukaryotes, and consequently they are suitable targets for protein-specific drugs.
Keywords: DEAD-box; Ded1/DDX3; Leishmania; RNA helicase; Saccharomyces cerevisiae; Trypanosoma brucei; leishmaniasis; trypanosomatid.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
The Ded1/DDX3 subfamily of DEAD-box RNA helicases.Crit Rev Biochem Mol Biol. 2014 Jul-Aug;49(4):343-60. doi: 10.3109/10409238.2014.931339. Crit Rev Biochem Mol Biol. 2014. PMID: 25039764 Review.
-
Gene duplication in trypanosomatids - two DED1 paralogs are functionally redundant and differentially expressed during the life cycle.Mol Biochem Parasitol. 2012 Oct;185(2):127-36. doi: 10.1016/j.molbiopara.2012.08.001. Epub 2012 Aug 14. Mol Biochem Parasitol. 2012. PMID: 22910033
-
The RNA Helicase Ded1 from Yeast Is Associated with the Signal Recognition Particle and Is Regulated by SRP21.Molecules. 2024 Jun 20;29(12):2944. doi: 10.3390/molecules29122944. Molecules. 2024. PMID: 38931009 Free PMC article.
-
Medulloblastoma-associated mutations in the DEAD-box RNA helicase DDX3X/DED1 cause specific defects in translation.J Biol Chem. 2021 Jan-Jun;296:100296. doi: 10.1016/j.jbc.2021.100296. Epub 2021 Jan 16. J Biol Chem. 2021. PMID: 33460649 Free PMC article.
-
The DDX3 subfamily of the DEAD box helicases: divergent roles as unveiled by studying different organisms and in vitro assays.Curr Med Chem. 2007;14(23):2517-25. doi: 10.2174/092986707782023677. Curr Med Chem. 2007. PMID: 17979704 Review.
Cited by
-
Distinct mRNA and protein interactomes highlight functional differentiation of major eIF4F-like complexes from Trypanosoma brucei.Front Mol Biosci. 2022 Oct 7;9:971811. doi: 10.3389/fmolb.2022.971811. eCollection 2022. Front Mol Biosci. 2022. PMID: 36275617 Free PMC article.
-
Enzymatic and Molecular Characterization of Anti-Leishmania Molecules That Differently Target Leishmania and Mammalian eIF4A Proteins, LieIF4A and eIF4AMus.Molecules. 2022 Sep 10;27(18):5890. doi: 10.3390/molecules27185890. Molecules. 2022. PMID: 36144626 Free PMC article.
-
Genetic depletion of the RNA helicase DDX3 leads to impaired elongation of translating ribosomes triggering co-translational quality control of newly synthesized polypeptides.Nucleic Acids Res. 2021 Sep 20;49(16):9459-9478. doi: 10.1093/nar/gkab667. Nucleic Acids Res. 2021. PMID: 34358325 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

