A diagnostic host response biosignature for COVID-19 from RNA profiling of nasal swabs and blood
- PMID: 33536218
- PMCID: PMC7857687
- DOI: 10.1126/sciadv.abe5984
A diagnostic host response biosignature for COVID-19 from RNA profiling of nasal swabs and blood
Abstract
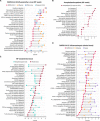
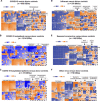
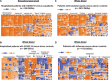
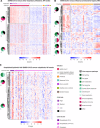
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes coronavirus disease-19 (COVID-19), has emerged as the cause of a global pandemic. We used RNA sequencing to analyze 286 nasopharyngeal (NP) swab and 53 whole-blood (WB) samples from 333 patients with COVID-19 and controls. Overall, a muted immune response was observed in COVID-19 relative to other infections (influenza, other seasonal coronaviruses, and bacterial sepsis), with paradoxical down-regulation of several key differentially expressed genes. Hospitalized patients and outpatients exhibited up-regulation of interferon-associated pathways, although heightened and more robust inflammatory responses were observed in hospitalized patients with more clinically severe illness. Two-layer machine learning-based host classifiers consisting of complete (>1000 genes), medium (<100), and small (<20) gene biomarker panels identified COVID-19 disease with 85.1-86.5% accuracy when benchmarked using an independent test set. SARS-CoV-2 infection has a distinct biosignature that differs between NP swabs and WB and can be leveraged for COVID-19 diagnosis.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures


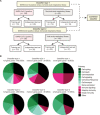

References
-
- Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D. S. C., Du B., Li L.-J., Zeng G., Yuen K.-Y., Chen R.-C., Tang C.-L., Wang T., Chen P.-Y., Xiang J., Li S.-Y., Wang J.-L., Liang Z.-J., Peng Y.-X., Wei L., Liu Y., Hu Y.-H., Peng P., Wang J.-M., Liu J.-Y., Chen Z., Li G., Zheng Z.-J., Qiu S.-Q., Luo J., Ye C.-J., Zhu S.-Y., Zhong N.-S.; China Medical Treatment Expert Group for Covid-19 , Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 382, 1708–1720 (2020). - PMC - PubMed
-
- Russell T. W., Hellewell J., Jarvis C. I., van Zandvoort K., Abbott S., Ratnayake R.; Cmmid Covid-Working Group, Flasche S., Eggo R. M., Edmunds W. J., Kucharski A. J., Estimating the infection and case fatality ratio for coronavirus disease (COVID-19) using age-adjusted data from the outbreak on the Diamond Princess cruise ship, February 2020. Euro Surveill. 25, 2000256 (2020). - PMC - PubMed
-
- Verity R., Okell L. C., Dorigatti I., Winskill P., Whittaker C., Imai N., Cuomo-Dannenburg G., Thompson H., Walker P. G. T., Fu H., Dighe A., Griffin J. T., Baguelin M., Bhatia S., Boonyasiri A., Cori A., Cucunuba Z., FitzJohn R., Gaythorpe K., Green W., Hamlet A., Hinsley W., Laydon D., Nedjati-Gilani G., Riley S., van Elsland S., Volz E., Wang H., Wang Y., Xi X., Donnelly C. A., Ghani A. C., Ferguson N. M., Estimates of the severity of coronavirus disease 2019: A model-based analysis. Lancet Infect. Dis. 20, 669–677 (2020). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

