Structural comparison of GLUT1 to GLUT3 reveal transport regulation mechanism in sugar porter family
- PMID: 33536238
- PMCID: PMC7898563
- DOI: 10.26508/lsa.202000858
Structural comparison of GLUT1 to GLUT3 reveal transport regulation mechanism in sugar porter family
Abstract
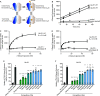
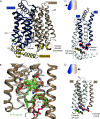
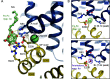
The human glucose transporters GLUT1 and GLUT3 have a central role in glucose uptake as canonical members of the Sugar Porter (SP) family. GLUT1 and GLUT3 share a fully conserved substrate-binding site with identical substrate coordination, but differ significantly in transport affinity in line with their physiological function. Here, we present a 2.4 Å crystal structure of GLUT1 in an inward open conformation and compare it with GLUT3 using both structural and functional data. Our work shows that interactions between a cytosolic "SP motif" and a conserved "A motif" stabilize the outward conformational state and increases substrate apparent affinity. Furthermore, we identify a previously undescribed Cl- ion site in GLUT1 and an endofacial lipid/glucose binding site which modulate GLUT kinetics. The results provide a possible explanation for the difference between GLUT1 and GLUT3 glucose affinity, imply a general model for the kinetic regulation in GLUTs and suggest a physiological function for the defining SP sequence motif in the SP family.
© 2021 Custódio et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







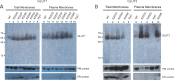



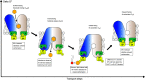
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. 10.1107/s0907444909052925 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
