Investigation of Heterochromatin Protein 1 Function in the Malaria Parasite Plasmodium falciparum Using a Conditional Domain Deletion and Swapping Approach
- PMID: 33536327
- PMCID: PMC7860992
- DOI: 10.1128/mSphere.01220-20
Investigation of Heterochromatin Protein 1 Function in the Malaria Parasite Plasmodium falciparum Using a Conditional Domain Deletion and Swapping Approach
Abstract
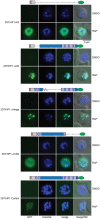
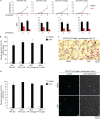
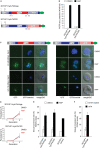
The human malaria parasite Plasmodium falciparum encodes a single ortholog of heterochromatin protein 1 (PfHP1) that plays a crucial role in the epigenetic regulation of various survival-related processes. PfHP1 is essential for parasite proliferation and the heritable silencing of genes linked to antigenic variation, host cell invasion, and sexual conversion. Here, we employed CRISPR/Cas9-mediated genome editing combined with the DiCre/loxP system to investigate how the PfHP1 chromodomain (CD), hinge domain, and chromoshadow domain (CSD) contribute to overall PfHP1 function. We show that the 76 C-terminal residues are responsible for targeting PfHP1 to the nucleus. Furthermore, we reveal that each of the three functional domains of PfHP1 are required for heterochromatin formation, gene silencing, and mitotic parasite proliferation. Finally, we discovered that the hinge domain and CSD of HP1 are functionally conserved between P. falciparum and P. berghei, a related malaria parasite infecting rodents. In summary, our study provides new insights into PfHP1 function and offers a tool for further studies on epigenetic regulation and life cycle decision in malaria parasites.IMPORTANCE Malaria is caused by unicellular Plasmodium species parasites that repeatedly invade and replicate inside red blood cells. Some blood-stage parasites exit the cell cycle and differentiate into gametocytes that are essential for malaria transmission via the mosquito vector. Epigenetic control mechanisms allow the parasites to alter the expression of surface antigens and to balance the switch between parasite multiplication and gametocyte production. These processes are crucial to establish chronic infection and optimize parasite transmission. Here, we performed a mutational analysis of heterochromatin protein 1 (HP1) in P. falciparum We demonstrate that all three domains of this protein are indispensable for the proper function of HP1 in parasite multiplication, heterochromatin formation, and gene silencing. Moreover, expression of chimeric proteins revealed the functional conservation of HP1 proteins between different Plasmodium species. These results provide new insight into the function and evolution of HP1 as an essential epigenetic regulator of parasite survival.
Keywords: CRISPR/Cas9; DiCre; HP1; Plasmodium falciparum; epigenetics; heterochromatin; malaria.
Copyright © 2021 Bui et al.
Figures

References
-
- Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SC. 1990. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci U S A 87:9923–9927. doi:10.1073/pnas.87.24.9923. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

