Population structure, genetic diversity and genomic selection signatures among a Brazilian common bean germplasm
- PMID: 33536468
- PMCID: PMC7859210
- DOI: 10.1038/s41598-021-82437-4
Population structure, genetic diversity and genomic selection signatures among a Brazilian common bean germplasm
Abstract
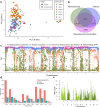
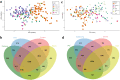
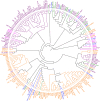
Brazil is the world's largest producer of common bean. Knowledge of the genetic diversity and relatedness of accessions adapted to Brazilian conditions is of great importance for the conservation of germplasm and for directing breeding programs aimed at the development of new cultivars. In this context, the objective of this study was to analyze the genetic diversity, population structure, and linkage disequilibrium (LD) of a diversity panel consisting of 219 common bean accessions, most of which belonging to the Mesoamerican gene pool. Genotyping by sequencing (GBS) of these accessions allowed the identification of 49,817 SNPs with minor allele frequency > 0.05. Of these, 17,149 and 12,876 were exclusive to the Mesoamerican and Andean pools, respectively, and 11,805 SNPs could differentiate the two gene pools. Further the separation according to the gene pool, bayesian analysis of the population structure showed a subdivision of the Mesoamerican accessions based on the origin and color of the seed tegument. LD analysis revealed the occurrence of long linkage blocks and low LD decay with physical distance between SNPs (LD half decay in 249 kb, corrected for population structure and relatedness). The GBS technique could effectively characterize the Brazilian common bean germplasms, and the diversity panel used in this study may be of great use in future genome-wide association studies.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Broughton WJ, et al. Beans (Phaseolus spp.)—model food legumes. Plant Soil. 2003;252:55–128. doi: 10.1023/A:1024146710611. - DOI
-
- FAO, F. and A. O. FAOSTAT: FAO Statistical Databases. http://www.fao.org/faostat/ (2020).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

