Salicylic acid stabilizes Staphylococcus aureus biofilm by impairing the agr quorum-sensing system
- PMID: 33536503
- PMCID: PMC7858585
- DOI: 10.1038/s41598-021-82308-y
Salicylic acid stabilizes Staphylococcus aureus biofilm by impairing the agr quorum-sensing system
Abstract
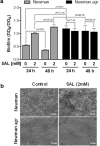
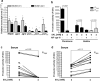
Salicylic acid (SAL) has recently been shown to induce biofilm formation in Staphylococcus aureus and to affect the expression of virulence factors. This study was aimed to investigate the effect of SAL on the regulatory agr system and its impact on S. aureus biofilm formation. The agr quorum-sensing system, which is a central regulator in S. aureus pathogenicity, plays a pivotal role in the dispersal of S. aureus mature biofilms and contributes to the creation of new colonization sites. Here, we demonstrate that SAL impairs biofilm dispersal by interfering with agr expression. As revealed by our work, protease and surfactant molecule production is diminished, and bacterial cell autolysis is also negatively affected by SAL. Furthermore, as a consequence of SAL treatment, the S. aureus biofilm matrix revealed the lack of extracellular DNA. In silico docking and simulation of molecular dynamics provided evidence for a potential interaction of AgrA and SAL, resulting in reduced activity of the agr system. In conclusion, SAL stabilized the mature S. aureus biofilms, which may prevent bacterial cell dissemination. However, it may foster the establishment of infections locally and consequently increase bacterial persistence leading to therapeutic failure.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Quercetin combined with ciprofloxacin and gentamicin inhibits biofilm formation and virulence in Staphylococcus aureus.Microb Pathog. 2025 Mar;200:107297. doi: 10.1016/j.micpath.2025.107297. Epub 2025 Jan 13. Microb Pathog. 2025. PMID: 39814109
-
Quorum sensing mediated attenuation of biofilm formation and virulence traits in Staphylococcus aureus by trigonelline.Microb Pathog. 2025 Aug;205:107731. doi: 10.1016/j.micpath.2025.107731. Epub 2025 May 21. Microb Pathog. 2025. PMID: 40409430
-
Luteolin attenuates the pathogenesis of Staphylococcus aureus by interfering with the agr system.Microb Pathog. 2022 Apr;165:105496. doi: 10.1016/j.micpath.2022.105496. Epub 2022 Mar 21. Microb Pathog. 2022. PMID: 35331848
-
Quorum sensing-mediated regulation of staphylococcal virulence and antibiotic resistance.Future Microbiol. 2014;9(5):669-81. doi: 10.2217/fmb.14.31. Future Microbiol. 2014. PMID: 24957093 Review.
-
What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia?Trends Microbiol. 2014 Dec;22(12):676-85. doi: 10.1016/j.tim.2014.09.002. Epub 2014 Oct 6. Trends Microbiol. 2014. PMID: 25300477 Review.
Cited by
-
Rational construction of genome-reduced Burkholderiales chassis facilitates efficient heterologous production of natural products from proteobacteria.Nat Commun. 2021 Jul 23;12(1):4347. doi: 10.1038/s41467-021-24645-0. Nat Commun. 2021. PMID: 34301933 Free PMC article.
-
Baicalein Inhibits the Staphylococcus aureus Biofilm and the LuxS/AI-2 System in vitro.Infect Drug Resist. 2023 May 9;16:2861-2882. doi: 10.2147/IDR.S406243. eCollection 2023. Infect Drug Resist. 2023. PMID: 37193303 Free PMC article.
-
In-depth characterization of accessory gene regulator loci and associated virulence factors in tcdA+B+ Clostridioides difficile isolates.Curr Res Microb Sci. 2025 Jul 1;9:100435. doi: 10.1016/j.crmicr.2025.100435. eCollection 2025. Curr Res Microb Sci. 2025. PMID: 40686843 Free PMC article.
-
Antimicrobial and anti-biofilm activity of Polygonum chinense L.aqueous extract against Staphylococcus aureus.Sci Rep. 2022 Dec 20;12(1):21988. doi: 10.1038/s41598-022-26399-1. Sci Rep. 2022. PMID: 36539472 Free PMC article.
-
N-Salicyl-AAn-picolamide Foldameric Peptides Exhibit Quorum Sensing Inhibition of Pseudomonas aeruginosa (PA14).ACS Omega. 2023 Aug 8;8(33):30349-30358. doi: 10.1021/acsomega.3c03404. eCollection 2023 Aug 22. ACS Omega. 2023. PMID: 37636905 Free PMC article.
References
-
- Yadav MK, Song JJ, Singh BP, Vidal JE. Microbial biofilms and human disease: a concise review. In: Yadav MK, Singh BP, editors. New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Biofilms. Ch 1. Amsterdam: Elsevier; 2020. pp. 1–13.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

