Elevated NSD3 histone methylation activity drives squamous cell lung cancer
- PMID: 33536620
- PMCID: PMC7895461
- DOI: 10.1038/s41586-020-03170-y
Elevated NSD3 histone methylation activity drives squamous cell lung cancer
Abstract
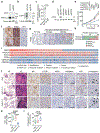
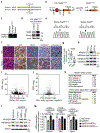
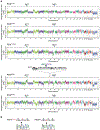
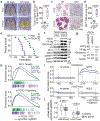
Amplification of chromosomal region 8p11-12 is a common genetic alteration that has been implicated in the aetiology of lung squamous cell carcinoma (LUSC)1-3. The FGFR1 gene is the main candidate driver of tumorigenesis within this region4. However, clinical trials evaluating FGFR1 inhibition as a targeted therapy have been unsuccessful5. Here we identify the histone H3 lysine 36 (H3K36) methyltransferase NSD3, the gene for which is located in the 8p11-12 amplicon, as a key regulator of LUSC tumorigenesis. In contrast to other 8p11-12 candidate LUSC drivers, increased expression of NSD3 correlated strongly with its gene amplification. Ablation of NSD3, but not of FGFR1, attenuated tumour growth and extended survival in a mouse model of LUSC. We identify an LUSC-associated variant NSD3(T1232A) that shows increased catalytic activity for dimethylation of H3K36 (H3K36me2) in vitro and in vivo. Structural dynamic analyses revealed that the T1232A substitution elicited localized mobility changes throughout the catalytic domain of NSD3 to relieve auto-inhibition and to increase accessibility of the H3 substrate. Expression of NSD3(T1232A) in vivo accelerated tumorigenesis and decreased overall survival in mouse models of LUSC. Pathological generation of H3K36me2 by NSD3(T1232A) reprograms the chromatin landscape to promote oncogenic gene expression signatures. Furthermore, NSD3, in a manner dependent on its catalytic activity, promoted transformation in human tracheobronchial cells and growth of xenografted human LUSC cell lines with amplification of 8p11-12. Depletion of NSD3 in patient-derived xenografts from primary LUSCs containing NSD3 amplification or the NSD3(T1232A)-encoding variant attenuated neoplastic growth in mice. Finally, NSD3-regulated LUSC-derived xenografts were hypersensitive to bromodomain inhibition. Thus, our work identifies NSD3 as a principal 8p11-12 amplicon-associated oncogenic driver in LUSC, and suggests that NSD3-dependency renders LUSC therapeutically vulnerable to bromodomain inhibition.
Figures










Comment in
-
An epigenetic tipping point in cancer comes under the microscope.Nature. 2021 Feb;590(7846):399-400. doi: 10.1038/d41586-021-00002-5. Nature. 2021. PMID: 33536596 No abstract available.
-
Squamous cell lung cancer changes driver.Nat Rev Drug Discov. 2021 Mar;20(3):177. doi: 10.1038/d41573-021-00028-4. Nat Rev Drug Discov. 2021. PMID: 33568801 No abstract available.
-
A driver gene hiding in plain sight.Nat Rev Cancer. 2021 Apr;21(4):213. doi: 10.1038/s41568-021-00341-5. Nat Rev Cancer. 2021. PMID: 33608665 No abstract available.
References
-
- Balsara BR et al. Comparative genomic hybridization analysis detects frequent, often high-level, overrepresentation of DNA sequences at 3q, 5p, 7p, and 8q in human non-small cell lung carcinomas. Cancer Res 57, 2116–2120 (1997). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

