Skin-resident innate lymphoid cells converge on a pathogenic effector state
- PMID: 33536623
- PMCID: PMC8336632
- DOI: 10.1038/s41586-021-03188-w
Skin-resident innate lymphoid cells converge on a pathogenic effector state
Abstract
Tissue-resident innate lymphoid cells (ILCs) help sustain barrier function and respond to local signals. ILCs are traditionally classified as ILC1, ILC2 or ILC3 on the basis of their expression of specific transcription factors and cytokines1. In the skin, disease-specific production of ILC3-associated cytokines interleukin (IL)-17 and IL-22 in response to IL-23 signalling contributes to dermal inflammation in psoriasis. However, it is not known whether this response is initiated by pre-committed ILCs or by cell-state transitions. Here we show that the induction of psoriasis in mice by IL-23 or imiquimod reconfigures a spectrum of skin ILCs, which converge on a pathogenic ILC3-like state. Tissue-resident ILCs were necessary and sufficient, in the absence of circulatory ILCs, to drive pathology. Single-cell RNA-sequencing (scRNA-seq) profiles of skin ILCs along a time course of psoriatic inflammation formed a dense transcriptional continuum-even at steady state-reflecting fluid ILC states, including a naive or quiescent-like state and an ILC2 effector state. Upon disease induction, the continuum shifted rapidly to span a mixed, ILC3-like subset also expressing cytokines characteristic of ILC2s, which we inferred as arising through multiple trajectories. We confirmed the transition potential of quiescent-like and ILC2 states using in vitro experiments, single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) and in vivo fate mapping. Our results highlight the range and flexibility of skin ILC responses, suggesting that immune activities primed in healthy tissues dynamically adapt to provocations and, left unchecked, drive pathological remodelling.
Conflict of interest statement
Figures
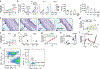










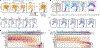
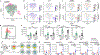
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

