4-Hydroxyphenylpyruvate Dioxygenase-Like Protein Promotes Pancreatic Cancer Cell Progression and Is Associated With Glutamine-Mediated Redox Balance
- PMID: 33537239
- PMCID: PMC7848781
- DOI: 10.3389/fonc.2020.617190
4-Hydroxyphenylpyruvate Dioxygenase-Like Protein Promotes Pancreatic Cancer Cell Progression and Is Associated With Glutamine-Mediated Redox Balance
Abstract
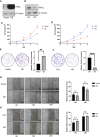
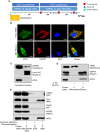
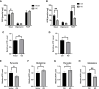
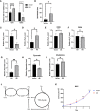
Tumor cells develop a series of metabolic reprogramming mechanisms to meet the metabolic needs for tumor progression. As metabolic hubs in cells, mitochondria play a significant role in this process, including energy production, biosynthesis, and redox hemostasis. In this study, we show that 4-hydroxyphenylpyruvate dioxygenase-like protein (HPDL), a previously uncharacterized protein, is positively associated with the development of pancreatic ductal adenocarcinoma (PDAC) and disease prognosis. We found that overexpression of HPDL in PDAC cells promotes tumorigenesis in vitro, whereas knockdown of HPDL inhibits cell proliferation and colony formation. Mechanistically, we found that HPDL is a mitochondrial intermembrane space localized protein that positively regulates mitochondrial bioenergetic processes and adenosine triphosphate (ATP) generation in a glutamine dependent manner. Our results further reveal that HPDL protects cells from oxidative stress by reprogramming the metabolic profile of PDAC cells toward glutamine metabolism. In short, we conclude that HPDL promotes PDAC likely through its effects on glutamine metabolism and redox balance.
Keywords: glutamine; metabolic reprogramming; mitochondria; pancreatic ductal adenocarcinoma; redox balance.
Copyright © 2021 Ye, Wei, Liao, Chen, Li, Chen, Yang, Zhao, Sun, Pan, Chen, He, Lyu and Fang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

