Mutalyzer 2: next generation HGVS nomenclature checker
- PMID: 33538839
- PMCID: PMC8479679
- DOI: 10.1093/bioinformatics/btab051
Mutalyzer 2: next generation HGVS nomenclature checker
Abstract
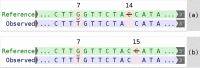
Motivation: Unambiguous variant descriptions are of utmost importance in clinical genetic diagnostics, scientific literature and genetic databases. The Human Genome Variation Society (HGVS) publishes a comprehensive set of guidelines on how variants should be correctly and unambiguously described. We present the implementation of the Mutalyzer 2 tool suite, designed to automatically apply the HGVS guidelines so users do not have to deal with the HGVS intricacies explicitly to check and correct their variant descriptions.
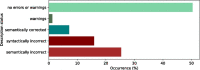
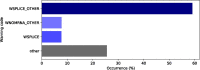
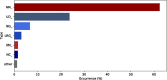
Results: Mutalyzer is profusely used by the community, having processed over 133 million descriptions since its launch. Over a five year period, Mutalyzer reported a correct input in ∼50% of cases. In 41% of the cases either a syntactic or semantic error was identified and for ∼7% of cases, Mutalyzer was able to automatically correct the description.
Availability and implementation: Mutalyzer is an Open Source project under the GNU Affero General Public License. The source code is available on GitHub (https://github.com/mutalyzer/mutalyzer) and a running instance is available at: https://mutalyzer.nl.
© The Author(s) 2021. Published by Oxford University Press.
Figures



References
-
- Ad Hoc Committee on Mutation Nomenclature (1996) Update on nomenclature for human gene mutations. Hum. Mutat., 8, 197–202. - PubMed
-
- Antonarakis S. et al. (1998) Recommendations for a nomenclature system for human gene mutations. Hum. Mutat., 11, 1–3. - PubMed
-
- Beaudet A., Tsui L.-C. (1993) A suggested nomenclature for designating mutations. Hum. Mutat., 2, 245–248. - PubMed
-
- Box D. et al. (2000) Simple Object Access Protocol (SOAP) 1.1. https://www.w3.org/TR/2000/NOTE-SOAP-20000508/.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

