ENAM gene associated with T classification and inhibits proliferation in renal clear cell carcinoma
- PMID: 33539322
- PMCID: PMC7993715
- DOI: 10.18632/aging.202558
ENAM gene associated with T classification and inhibits proliferation in renal clear cell carcinoma
Abstract
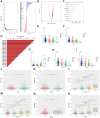
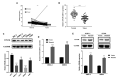
The potential involvement of T classification-related genes in renal clear cell carcinoma (ccRCC) must be further explored. Public data were obtained from The Cancer Genome Atlas (TCGA) database. An overall survival (OS) predictive model was developed and validated (TCGA train, 5 years, AUC = 0.73, 3 years, AUC = 0.73, 1 year, AUC = 0.76; TCGA test, 5 years, AUC = 0.74, 3 years, AUC = 0.65, 1 year, AUC = 0.73; TCGA all, 5 years, AUC = 0.72, 3 years, AUC = 0.71, 1 year, AUC = 0.75). Finally, ENAM was selected for further analysis. In vitro experiment indicated that ENMA is downregulated in ccRCC, and its knockdown could promote proliferation in two cancer cell lines (OSRC-2 and SW839). Immune infiltration analysis revealed that ENAM could remarkably increase the content of cytotoxic cells, NK CD56 cells, NK cells and CD8+ T cells in the tumor immune microenvironment, which may be one reason for its tumor-inhibiting effect. In summary, ENAM may suppress cell proliferation in ccRCC and can be used as a potential reference value for the relief and immunotherapy of ccRCC.
Keywords: ENAM; T classification; renal cell carcinoma.
Conflict of interest statement
Figures





References
-
- Gremel G, Djureinovic D, Niinivirta M, Laird A, Ljungqvist O, Johannesson H, Bergman J, Edqvist PH, Navani S, Khan N, Patil T, Sivertsson Å, Uhlén M, et al. A systematic search strategy identifies cubilin as independent prognostic marker for renal cell carcinoma. BMC Cancer. 2017; 17:9. 10.1186/s12885-016-3030-6 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

