OLIG2 maintenance is not essential for diffuse intrinsic pontine glioma cell line growth but regulates tumor phenotypes
- PMID: 33539525
- PMCID: PMC8661399
- DOI: 10.1093/neuonc/noab016
OLIG2 maintenance is not essential for diffuse intrinsic pontine glioma cell line growth but regulates tumor phenotypes
Abstract
Background: Diffuse intrinsic pontine glioma (DIPG) is a pediatric lethal high-grade brainstem glioma with no effective therapies. OLIG2 (oligodendrocyte transcription factor 2) was reported to be critical for the growth of a DIPG cell line CCHMC-DIPG-1. Surprisingly, we found that the CCHMC-DIPG-1 cells express little OLIG2 and exhibit a mesenchymal phenotype, which raised a question regarding the role of OLIG2 in the growth of DIPG cells.
Methods: We evaluated the function of OLIG2 in different DIPG cell lines through molecular and genetic approaches and performed transcriptomic and genomic landscape profiling including whole-genome bisulfite sequencing, RNA-seq, ATAC-seq, and ChIP-seq. shRNA-mediated knockdown and CRISPR-Cas9-mediated knockout approaches were utilized to assess OLIG2 functions in DIPG cell growth.
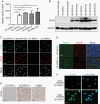
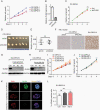
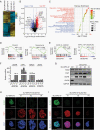
Results: We found that DIPG cells are phenotypically heterogeneous and exhibit the characteristics of distinct malignant gliomas including proneural, classical, and mesenchymal subtypes. OLIG2 knockdown did not impact the growth of CCHMC-DIPG-1 cells, wherein OLIG2 is epigenetically silenced. Moreover, OLIG2 deletion did not substantially impair OLIG2-expressing proneural-like DIPG growth but led to an upregulation of HIPPO-YAP1 and epidermal growth factor receptor (EGFR) signaling and a tumor phenotype shift. Targeting HIPPO-YAP1 and EGFR signaling in OLIG2-deficient DIPG cells inhibited tumor cell growth.
Conclusions: Our data indicate that OLIG2 is dispensable for DIPG growth but regulates the phenotypic switch of DIPG tumor cells. OLIG2 downregulation leads to deregulation of adaptive YAP1 and EGFR signaling. Targeting YAP1 and EGFR pathways inhibits the growth of OLIG2-deficient DIPG cells, pointing to a therapeutic potential by targeting adaptive signaling to treat DIPG tumors with nominal OLIG2 expression.
Keywords: OLIG2; YAP1 and EGFR signaling; diffuse intrinsic pontine glioma (DIPG); distinct DIPG tumor phenotypes; genomic landscapes.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

