Polycystic ovary syndrome is transmitted via a transgenerational epigenetic process
- PMID: 33539777
- PMCID: PMC7928942
- DOI: 10.1016/j.cmet.2021.01.004
Polycystic ovary syndrome is transmitted via a transgenerational epigenetic process
Abstract
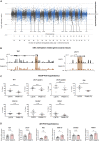
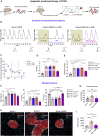
Polycystic ovary syndrome (PCOS) is the most common reproductive and metabolic disorder affecting women of reproductive age. PCOS has a strong heritable component, but its pathogenesis has been unclear. Here, we performed RNA sequencing and genome-wide DNA methylation profiling of ovarian tissue from control and third-generation PCOS-like mice. We found that DNA hypomethylation regulates key genes associated with PCOS and that several of the differentially methylated genes are also altered in blood samples from women with PCOS compared with healthy controls. Based on this insight, we treated the PCOS mouse model with the methyl group donor S-adenosylmethionine and found that it corrected their transcriptomic, neuroendocrine, and metabolic defects. These findings show that the transmission of PCOS traits to future generations occurs via an altered landscape of DNA methylation and propose methylome markers as a possible diagnostic landmark for the condition, while also identifying potential candidates for epigenetic-based therapy.
Keywords: AMH; PCOS; developmental programming; epigenetics; fertility; inheritance; metabolic disorder; methylation; neuroendocrine; transgenerational.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests P.G., N.E.H.M., I.P., A.-L.B., and V.P. disclose that they are inventors of a submitted patent application by the INSERM (Institut National de la Santé et de la Recherche Médicale) covering methods and kits for diagnostic and treatment of PCOS. All other authors do not have competing interests.
Figures
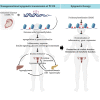
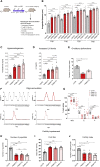
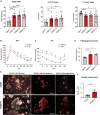

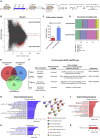

Comment in
-
Passing on PCOS: new insights into its epigenetic transmission.Cell Metab. 2021 Mar 2;33(3):463-466. doi: 10.1016/j.cmet.2021.02.008. Cell Metab. 2021. PMID: 33657389 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

