Dynamics of a Dual SARS-CoV-2 Lineage Co-Infection on a Prolonged Viral Shedding COVID-19 Case: Insights into Clinical Severity and Disease Duration
- PMID: 33540596
- PMCID: PMC7912897
- DOI: 10.3390/microorganisms9020300
Dynamics of a Dual SARS-CoV-2 Lineage Co-Infection on a Prolonged Viral Shedding COVID-19 Case: Insights into Clinical Severity and Disease Duration
Abstract
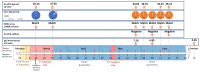
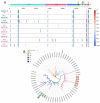
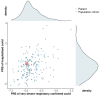
A few molecularly proven severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) cases of symptomatic reinfection are currently known worldwide, with a resolved first infection followed by a second infection after a 48 to 142-day intervening period. We report a multiple-component study of a clinically severe and prolonged viral shedding coronavirus disease 2019 (COVID-19) case in a 17-year-old Portuguese female. She had two hospitalizations, a total of 19 RT-PCR tests, mostly positive, and criteria for releasing from home isolation at the end of 97 days. The viral genome was sequenced in seven serial samples and in the diagnostic sample from her infected mother. A human genome-wide array (>900 K) was screened on the seven samples, and in vitro culture was conducted on isolates from three late samples. The patient had co-infection by two SARS-CoV-2 lineages, which were affiliated in distinct clades and diverging by six variants. The 20A lineage was absolute at the diagnosis (shared with the patient's mother), but nine days later, the 20B lineage had 3% frequency, and two months later, the 20B lineage had 100% frequency. The 900 K profiles confirmed the identity of the patient in the serial samples, and they allowed us to infer that she had polygenic risk scores for hospitalization and severe respiratory disease within the normal distributions for a Portuguese population cohort. The early-on dynamic co-infection may have contributed to the severity of COVID-19 in this otherwise healthy young patient, and to her prolonged SARS-CoV-2 shedding profile.
Keywords: COVID-19 and SARS-CoV-2; co-infection; polygenic risk score; viral shedding; viral whole-genome sequencing.
Conflict of interest statement
No reported conflicts of interest.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

