Novakomyces olei sp. nov., the First Member of a Novel Taphrinomycotina Lineage
- PMID: 33540601
- PMCID: PMC7912804
- DOI: 10.3390/microorganisms9020301
Novakomyces olei sp. nov., the First Member of a Novel Taphrinomycotina Lineage
Abstract
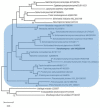
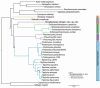
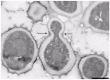
Taphrinomycotina is the smallest subphylum of the phylum Ascomycota. It is an assemblage of distantly related early diverging lineages of the phylum, comprising organisms with divergent morphology and ecology; however, phylogenomic analyses support its monophyly. In this study, we report the isolation of a yeast strain, which could not be assigned to any of the currently recognised five classes of Taphrinomycotina. The strain of the novel budding species was recovered from extra virgin olive oil and characterised phenotypically by standard methods. The ultrastructure of the cell wall was investigated by transmission electron microscopy. Comparisons of barcoding DNA sequences indicated that the investigated strain is not closely related to any known organism. Tentative phylogenetic placement was achieved by maximum-likelihood analysis of the D1/D2 domain of the nuclear LSU rRNA gene. The genome of the investigated strain was sequenced, assembled, and annotated. Phylogenomic analyses placed it next to the fission Schizosaccharomyces species. To accommodate the novel species, Novakomyces olei, a novel genus Novakomyces, a novel family Novakomycetaceae, a novel order Novakomycetales, and a novel class Novakomycetes is proposed as well. Functional analysis of genes missing in N. olei in comparison to Schizosaccharomyces pombe revealed that they are biased towards biosynthesis of complex organic molecules, regulation of mRNA, and the electron transport chain. Correlating the genome content and physiology among species of Taphrinomycotina revealed some discordance between pheno- and genotype. N. olei produced ascospores in axenic culture preceded by conjugation between two cells. We confirmed that N. olei is a primary homothallic species lacking genes for different mating types.
Keywords: functional gene analysis; genome sequence; novel class; novel yeast species; olive oil; phylogenomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Phylogenomic analyses support the monophyly of Taphrinomycotina, including Schizosaccharomyces fission yeasts.Mol Biol Evol. 2009 Jan;26(1):27-34. doi: 10.1093/molbev/msn221. Epub 2008 Oct 14. Mol Biol Evol. 2009. PMID: 18922765 Free PMC article.
-
Early diverging Ascomycota: phylogenetic divergence and related evolutionary enigmas.Mycologia. 2006 Nov-Dec;98(6):996-1005. doi: 10.3852/mycologia.98.6.996. Mycologia. 2006. PMID: 17486975
-
Saitoella coloradoensis sp. nov., a new species of the Ascomycota, subphylum Taphrinomycotina.Antonie Van Leeuwenhoek. 2012 May;101(4):795-802. doi: 10.1007/s10482-011-9694-7. Epub 2012 Jan 13. Antonie Van Leeuwenhoek. 2012. PMID: 22246589
-
Candida adriatica sp. nov. and Candida molendinolei sp. nov., two yeast species isolated from olive oil and its by-products.Int J Syst Evol Microbiol. 2012 Sep;62(Pt 9):2296-2302. doi: 10.1099/ijs.0.038794-0. Epub 2012 Jan 6. Int J Syst Evol Microbiol. 2012. PMID: 22228664
-
Tepidiphilus olei sp. nov., isolated from the production water of a water-flooded oil reservoir in PR China.Int J Syst Evol Microbiol. 2020 Jul;70(7):4364-4371. doi: 10.1099/ijsem.0.004297. Int J Syst Evol Microbiol. 2020. PMID: 32579101
Cited by
-
Detection and Identification of Food-Borne Yeasts: An Overview of the Relevant Methods and Their Evolution.Microorganisms. 2025 Apr 24;13(5):981. doi: 10.3390/microorganisms13050981. Microorganisms. 2025. PMID: 40431154 Free PMC article. Review.
-
A genome-informed higher rank classification of the biotechnologically important fungal subphylum Saccharomycotina.Stud Mycol. 2023 Jun;105:1-22. doi: 10.3114/sim.2023.105.01. Epub 2023 May 25. Stud Mycol. 2023. PMID: 38895705 Free PMC article.
-
Retracing the evolution of Pneumocystis species, with a focus on the human pathogen Pneumocystis jirovecii.Microbiol Mol Biol Rev. 2024 Jun 27;88(2):e0020222. doi: 10.1128/mmbr.00202-22. Epub 2024 Apr 8. Microbiol Mol Biol Rev. 2024. PMID: 38587383 Free PMC article. Review.
References
-
- Eriksson O.E., Winka K. Supraordinal taxa of Ascomycota. Myconet. 1997;1:1–16.
-
- Nishida H., Sugiyama J. Archiascomycetes: Detection of a major new lineage within the Ascomycota. Mycoscience. 1994;35:361–366. doi: 10.1007/BF02268506. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

