Cntn4, a risk gene for neuropsychiatric disorders, modulates hippocampal synaptic plasticity and behavior
- PMID: 33542194
- PMCID: PMC7862349
- DOI: 10.1038/s41398-021-01223-y
Cntn4, a risk gene for neuropsychiatric disorders, modulates hippocampal synaptic plasticity and behavior
Abstract
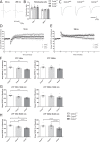
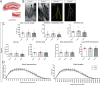
Neurodevelopmental and neuropsychiatric disorders, such as autism spectrum disorders (ASD), anorexia nervosa (AN), Alzheimer's disease (AD), and schizophrenia (SZ), are heterogeneous brain disorders with unknown etiology. Genome wide studies have revealed a wide variety of risk genes for these disorders, indicating a biological link between genetic signaling pathways and brain pathology. A unique risk gene is Contactin 4 (Cntn4), an Ig cell adhesion molecule (IgCAM) gene, which has been associated with several neuropsychiatric disorders including ASD, AN, AD, and SZ. Here, we investigated the Cntn4 gene knockout (KO) mouse model to determine whether memory dysfunction and altered brain plasticity, common neuropsychiatric symptoms, are affected by Cntn4 genetic disruption. For that purpose, we tested if Cntn4 genetic disruption affects CA1 synaptic transmission and the ability to induce LTP in hippocampal slices. Stimulation in CA1 striatum radiatum significantly decreased synaptic potentiation in slices of Cntn4 KO mice. Neuroanatomical analyses showed abnormal dendritic arborization and spines of hippocampal CA1 neurons. Short- and long-term recognition memory, spatial memory, and fear conditioning responses were also assessed. These behavioral studies showed increased contextual fear conditioning in heterozygous and homozygous KO mice, quantified by a gene-dose dependent increase in freezing response. In comparison to wild-type mice, Cntn4-deficient animals froze significantly longer and groomed more, indicative of increased stress responsiveness under these test conditions. Our electrophysiological, neuro-anatomical, and behavioral results in Cntn4 KO mice suggest that Cntn4 has important functions related to fear memory possibly in association with the neuronal morphological and synaptic plasticity changes in hippocampus CA1 neurons.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- World Health Organization. Depression and other common mental disorders: global health estimates. Geneva: World Health Organization; (2017). Licence: CC BY-NC-SA 3.0 IGO.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

