The interplay of UBE2T and Mule in regulating Wnt/β-catenin activation to promote hepatocellular carcinoma progression
- PMID: 33542213
- PMCID: PMC7862307
- DOI: 10.1038/s41419-021-03403-6
The interplay of UBE2T and Mule in regulating Wnt/β-catenin activation to promote hepatocellular carcinoma progression
Abstract
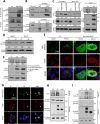
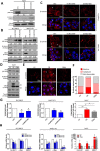
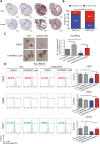
Emerging evidence indicates the role of cancer stem cells (CSCs) in tumor relapse and therapeutic resistance in patients with hepatocellular carcinoma (HCC). To identify novel targets against liver CSCs, an integrative analysis of publicly available datasets involving HCC clinical and stemness-related data was employed to select genes that play crucial roles in HCC via regulation of liver CSCs. We revealed an enrichment of an interstrand cross-link repair pathway, in which ubiquitin-conjugating enzyme E2 T (UBE2T) was the most significantly upregulated. Consistently, our data showed that UBE2T was upregulated in enriched liver CSC populations. Clinically, UBE2T overexpression in HCC was further confirmed at mRNA and protein levels and was correlated with advanced tumor stage and poor patient survival. UBE2T was found to be critically involved in the regulation of liver CSCs, as evidenced by increases in self-renewal, drug resistance, tumorigenicity, and metastasis abilities. Mule, an E3 ubiquitin ligase, was identified to be the direct protein binding partner of UBE2T. Rather than the canonical role of acting as a mediator to transfer ubiquitin to E3 ligases, UBE2T is surprisingly able to physically bind and regulate the protein expression of Mule via ubiquitination. Mule was found to directly degrade β-catenin protein, and UBE2T was found to mediate liver CSC functions through direct regulation of Mule-mediated β-catenin degradation; this effect was abolished when the E2 activity of UBE2T was impaired. In conclusion, we revealed a novel UBE2T/Mule/β-catenin signaling cascade that is involved in the regulation of liver CSCs, which provides an attractive potential therapeutic target for HCC.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Similar articles
-
UBE2T-mediated Akt ubiquitination and Akt/β-catenin activation promotes hepatocellular carcinoma development by increasing pyrimidine metabolism.Cell Death Dis. 2022 Feb 15;13(2):154. doi: 10.1038/s41419-022-04596-0. Cell Death Dis. 2022. PMID: 35169125 Free PMC article.
-
UBE2T-regulated H2AX monoubiquitination induces hepatocellular carcinoma radioresistance by facilitating CHK1 activation.J Exp Clin Cancer Res. 2020 Oct 21;39(1):222. doi: 10.1186/s13046-020-01734-4. J Exp Clin Cancer Res. 2020. PMID: 33087136 Free PMC article.
-
UBE2T promotes β-catenin nuclear translocation in hepatocellular carcinoma through MAPK/ERK-dependent activation.Mol Oncol. 2022 Apr;16(8):1694-1713. doi: 10.1002/1878-0261.13111. Epub 2022 Mar 16. Mol Oncol. 2022. PMID: 34614271 Free PMC article.
-
Targeting Wnt/β-catenin pathway in hepatocellular carcinoma treatment.World J Gastroenterol. 2016 Jan 14;22(2):823-32. doi: 10.3748/wjg.v22.i2.823. World J Gastroenterol. 2016. PMID: 26811628 Free PMC article. Review.
-
Unraveling the Role of Ubiquitin-Conjugating Enzyme UBE2T in Tumorigenesis: A Comprehensive Review.Cells. 2024 Dec 26;14(1):15. doi: 10.3390/cells14010015. Cells. 2024. PMID: 39791716 Free PMC article. Review.
Cited by
-
Cancer stem cells in hepatocellular carcinoma - from origin to clinical implications.Nat Rev Gastroenterol Hepatol. 2022 Jan;19(1):26-44. doi: 10.1038/s41575-021-00508-3. Epub 2021 Sep 9. Nat Rev Gastroenterol Hepatol. 2022. PMID: 34504325 Review.
-
UBE2S: A novel driver of HIF-1alpha-induced metabolic reprogramming in hepatocellular carcinoma: Editorial on "UBE2S promotes glycolysis in hepatocellular carcinoma by enhancing E3 enzyme-independent polyubiquitination of VHL".Clin Mol Hepatol. 2025 Jan;31(1):281-285. doi: 10.3350/cmh.2024.0568. Epub 2024 Jul 23. Clin Mol Hepatol. 2025. PMID: 39038959 Free PMC article. No abstract available.
-
UBE2T/STAT3 Signaling Promotes the Proliferation and Tumorigenesis in Retinoblastoma.Invest Ophthalmol Vis Sci. 2022 Aug 2;63(9):20. doi: 10.1167/iovs.63.9.20. Invest Ophthalmol Vis Sci. 2022. Retraction in: Invest Ophthalmol Vis Sci. 2025 Aug 1;66(11):56. doi: 10.1167/iovs.66.11.56. PMID: 35980647 Free PMC article. Retracted.
-
UBE2T promotes epithelial-mesenchymal transition and motility in oral cancer cells via induction of IL-6 expression.Oncol Lett. 2025 Aug 8;30(4):473. doi: 10.3892/ol.2025.15219. eCollection 2025 Oct. Oncol Lett. 2025. PMID: 40842469 Free PMC article.
-
Upregulation of ubiquitin-conjugating enzyme E2T (UBE2T) predicts poor prognosis and promotes hepatocellular carcinoma progression.Bioengineered. 2021 Dec;12(1):1530-1542. doi: 10.1080/21655979.2021.1918507. Bioengineered. 2021. PMID: 33934686 Free PMC article.
References
-
- Singh SK, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

