Identification of a solo acylhomoserine lactone synthase from the myxobacterium Archangium gephyra
- PMID: 33542315
- PMCID: PMC7862692
- DOI: 10.1038/s41598-021-82480-1
Identification of a solo acylhomoserine lactone synthase from the myxobacterium Archangium gephyra
Abstract
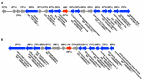
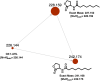
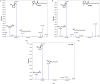
Considered a key taxon in soil and marine microbial communities, myxobacteria exist as coordinated swarms that utilize a combination of lytic enzymes and specialized metabolites to facilitate predation of microbes. This capacity to produce specialized metabolites and the associated abundance of biosynthetic pathways contained within their genomes have motivated continued drug discovery efforts from myxobacteria. Of all myxobacterial biosynthetic gene clusters deposited in the antiSMASH database, only one putative acylhomoserine lactone (AHL) synthase, agpI, was observed, in genome data from Archangium gephyra. Without an AHL receptor also apparent in the genome of A. gephyra, we sought to determine if AgpI was an uncommon example of an orphaned AHL synthase. Herein we report the bioinformatic assessment of AgpI and discovery of a second AHL synthase from Vitiosangium sp. During axenic cultivation conditions, no detectible AHL metabolites were observed in A. gephyra extracts. However, heterologous expression of each synthase in Escherichia coli provided detectible quantities of 3 AHL signals including 2 known AHLs, C8-AHL and C9-AHL. These results suggest that A. gephyra AHL production is dormant during axenic cultivation. The functional, orphaned AHL synthase, AgpI, is unique to A. gephyra, and its utility to the predatory myxobacterium remains unknown.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

