A recombinase polymerase amplification assay for rapid detection of rabies virus
- PMID: 33542337
- PMCID: PMC7862592
- DOI: 10.1038/s41598-021-82479-8
A recombinase polymerase amplification assay for rapid detection of rabies virus
Abstract
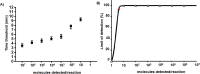
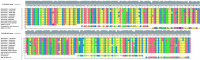
Rabies is a generally fatal encephalitis caused by a negative-sense single-stranded RNA lyssavirus transmitted to humans mainly from dog bite. Despite the recommendation by WHO and OIE to use the direct immunofluorescence test as standard method, molecular diagnostic assays like reverse transcription quantitative polymerase chain reaction (RT-qPCR) are increasing as a confirmatory method. However, both technologies are inaccessible in resource-limited settings. Moreover, the available point-of-need molecular assay is of poor detection limit for African strains. Herein, we developed a reverse transcription recombinase polymerase amplification (RT-RPA) assay as potential point-of-need diagnostic tool for rapid detection of various strains of rabies virus including locally isolated African strains. The sensitivity and specificity of the method was evaluated using a molecular RNA standard and different Rabies-related viruses belonging to the Rhabdoviridea family, respectively. The RABV-RPA performances were evaluated on isolates representative of the existing diversity and viral dilutions spiked in non-neural clinical specimen. The results were compared with RT-qPCR as a gold standard. The RABV-RPA detected down to 4 RNA molecules per reaction in 95% of the cases in less than 10 min. The RABV-RPA assay is highly specific as various RABV isolates were identified, but no amplification was observed for other member of the Rhabdoviridea family. The sample background did not affect the performance of the RABV-RPA as down to 11 RNA molecules were identified, which is similar to the RT-qPCR results. Our developed assay is suitable for use in low-resource settings as a promising alternative tool for ante-mortem rabies diagnosis in humans for facilitating timely control decisions.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Dietzschold B, et al. Pathogenesis of rabies. Curr. Top. Microbiol. Immunol. 2005;292:45–56. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

