Shared and Unique Features of Human Interferon-Beta and Interferon-Alpha Subtypes
- PMID: 33542718
- PMCID: PMC7850986
- DOI: 10.3389/fimmu.2020.605673
Shared and Unique Features of Human Interferon-Beta and Interferon-Alpha Subtypes
Abstract
Type I interferons (IFN-I) were first discovered as an antiviral factor by Isaacs and Lindenmann in 1957, but they are now known to also modulate innate and adaptive immunity and suppress proliferation of cancer cells. While much has been revealed about IFN-I, it remains a mystery as to why there are 16 different IFN-I gene products, including IFNβ, IFNω, and 12 subtypes of IFNα. Here, we discuss shared and unique aspects of these IFN-I in the context of their evolution, expression patterns, and signaling through their shared heterodimeric receptor. We propose that rather than investigating responses to individual IFN-I, these contexts can serve as an alternative approach toward investigating roles for IFNα subtypes. Finally, we review uses of IFNα and IFNβ as therapeutic agents to suppress chronic viral infections or to treat multiple sclerosis.
Keywords: human; interferon-alpha; interferon-beta; interferon-omega; primate; type I interferon.
Copyright © 2021 Wittling, Cahalan, Levenson and Rabin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
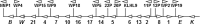

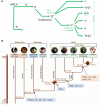
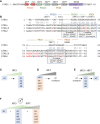
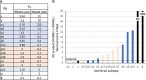

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

