Cartilage-like composition of keloid scar extracellular matrix suggests fibroblast mis-differentiation in disease
- PMID: 33543013
- PMCID: PMC7852214
- DOI: 10.1016/j.mbplus.2019.100016
Cartilage-like composition of keloid scar extracellular matrix suggests fibroblast mis-differentiation in disease
Abstract
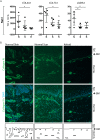
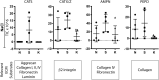
Following wound damage to the skin, the scarring spectrum is wide-ranging, from a manageable normal scar through to pathological keloids. The question remains whether these fibrotic lesions represent simply a quantitative extreme, or alternatively, whether they are qualitatively distinct. A three-way comparison of the extracellular matrix (ECM) composition of normal skin, normal scar and keloids was performed using quantitative discovery-based proteomics. This approach identified 40 proteins that were significantly altered in keloids compared to normal scars, and strikingly, 23 keloid-unique proteins. The major alterations in keloids, when functionally grouped, showed many changes in proteins involved in ECM assembly and fibrillogenesis, but also a keloid-associated loss of proteases, and a unique cartilage-like composition, which was also evident histologically. The presence of Aggrecan and Collagen II in keloids suggest greater plasticity and mis-differentiation of the constituent cells. This study characterises the ECM of both scar types to a depth previously underappreciated. This thorough molecular description of keloid lesions relative to normal scars is an essential step towards our understanding of this debilitating clinical problem, and how best to treat it.
Keywords: Differentiation; ECM; Fibrosis; Plasticity; Scar; Wound.
© 2019 The Authors.
Conflict of interest statement
The authors have declared that no conflict of interest exists.
Figures





References
-
- J. Shaw T., Kishi K., Mori R. Wound-associated skin fibrosis: mechanisms and treatments based on modulating the inflammatory response. Endocr Metab Immune Disord Drug Targets. 2010;10(4):320–330. - PubMed
-
- Lee J.Y., Yang C.C., Chao S.C., Wong T.W. Histopathological differential diagnosis of keloid and hypertrophic scar. Am. J. Dermatopathol. 2004;26(5):379–384. - PubMed
-
- Jumper N., Paus R., Bayat A. Functional histopathology of keloid disease. Histol. Histopathol. 2015;30(9):1033–1057. - PubMed
-
- Naitoh M., Kubota H., Ikeda M., Tanaka T., Shirane H., Suzuki S., Nagata K. Genes to Cells: Devoted to Molecular & Cellular Mechanisms. 10(11) 2005. Gene expression in human keloids is altered from dermal to chondrocytic and osteogenic lineage; pp. 1081–1091. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

