Cycles, sources, and sinks: Conceptualizing how phosphate balance modulates carbon flux using yeast metabolic networks
- PMID: 33544078
- PMCID: PMC7864628
- DOI: 10.7554/eLife.63341
Cycles, sources, and sinks: Conceptualizing how phosphate balance modulates carbon flux using yeast metabolic networks
Abstract
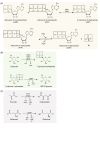
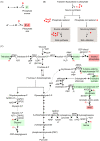
Phosphates are ubiquitous molecules that enable critical intracellular biochemical reactions. Therefore, cells have elaborate responses to phosphate limitation. Our understanding of long-term transcriptional responses to phosphate limitation is extensive. Contrastingly, a systems-level perspective presenting unifying biochemical concepts to interpret how phosphate balance is critically coupled to (and controls) metabolic information flow is missing. To conceptualize such processes, utilizing yeast metabolic networks we categorize phosphates utilized in metabolism into cycles, sources and sinks. Through this, we identify metabolic reactions leading to putative phosphate sources or sinks. With this conceptualization, we illustrate how mass action driven flux towards sources and sinks enable cells to manage phosphate availability during transient/immediate phosphate limitations. We thereby identify how intracellular phosphate availability will predictably alter specific nodes in carbon metabolism, and determine signature cellular metabolic states. Finally, we identify a need to understand intracellular phosphate pools, in order to address mechanisms of phosphate regulation and restoration.
Keywords: carbon metabolism; computational biology; gene expression; mass action; metabolic flux; metabolic networks; phosphate; systems biology.
© 2021, Gupta and Laxman.
Conflict of interest statement
RG, SL No competing interests declared
Figures

Similar articles
-
A tRNA modification balances carbon and nitrogen metabolism by regulating phosphate homeostasis.Elife. 2019 Jul 1;8:e44795. doi: 10.7554/eLife.44795. Elife. 2019. PMID: 31259691 Free PMC article.
-
Role of phosphate limitation and pyruvate decarboxylase in rewiring of the metabolic network for increasing flux towards isoprenoid pathway in a TATA binding protein mutant of Saccharomyces cerevisiae.Microb Cell Fact. 2018 Sep 21;17(1):152. doi: 10.1186/s12934-018-1000-1. Microb Cell Fact. 2018. PMID: 30241525 Free PMC article.
-
Heterologous phosphoketolase expression redirects flux towards acetate, perturbs sugar phosphate pools and increases respiratory demand in Saccharomyces cerevisiae.Microb Cell Fact. 2019 Feb 1;18(1):25. doi: 10.1186/s12934-019-1072-6. Microb Cell Fact. 2019. PMID: 30709397 Free PMC article.
-
A flux-sensing mechanism could regulate the switch between respiration and fermentation.FEMS Yeast Res. 2012 Mar;12(2):118-28. doi: 10.1111/j.1567-1364.2011.00767.x. Epub 2011 Dec 19. FEMS Yeast Res. 2012. PMID: 22129078 Review.
-
From network models to network responses: integration of thermodynamic and kinetic properties of yeast genome-scale metabolic networks.FEMS Yeast Res. 2012 Mar;12(2):129-43. doi: 10.1111/j.1567-1364.2011.00771.x. Epub 2011 Dec 19. FEMS Yeast Res. 2012. PMID: 22129227 Review.
Cited by
-
A DFT Study of Phosphate Ion Adsorption on Graphene Nanodots: Implications for Sensing.Sensors (Basel). 2023 Jun 16;23(12):5631. doi: 10.3390/s23125631. Sensors (Basel). 2023. PMID: 37420797 Free PMC article.
-
The deubiquitinase Ubp3/Usp10 constrains glucose-mediated mitochondrial repression via phosphate budgeting.Elife. 2024 Sep 26;12:RP90293. doi: 10.7554/eLife.90293. Elife. 2024. PMID: 39324403 Free PMC article.
-
Metabolic Consequences of Polyphosphate Synthesis and Imminent Phosphate Limitation.mBio. 2023 Jun 27;14(3):e0010223. doi: 10.1128/mbio.00102-23. Epub 2023 Apr 19. mBio. 2023. PMID: 37074217 Free PMC article.
-
Characterizing the regulatory effects of H2A.Z and SWR1-C on gene expression during hydroxyurea exposure in Saccharomyces cerevisiae.PLoS Genet. 2025 Jan 21;21(1):e1011566. doi: 10.1371/journal.pgen.1011566. eCollection 2025 Jan. PLoS Genet. 2025. PMID: 39836664 Free PMC article.
-
Ceramide Synthase 6 Maximizes p53 Function to Prevent Progeny Formation from Polyploid Giant Cancer Cells.Cancers (Basel). 2021 May 5;13(9):2212. doi: 10.3390/cancers13092212. Cancers (Basel). 2021. PMID: 34062962 Free PMC article.
References
-
- Ashihara H, Li X-NI, Ukaji T. Effect of inorganic phosphate on the biosynthesis of purine and pyrimidine nucleotides in Suspension-Cultured cells of Catharanthus roseus*. Annals of Botany. 1988;61:225–232. doi: 10.1093/oxfordjournals.aob.a087547. - DOI
-
- Auesukaree C, Tochio H, Shirakawa M, Kaneko Y, Harashima S. Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. Journal of Biological Chemistry. 2005;280:25127–25133. doi: 10.1074/jbc.M414579200. - DOI - PubMed
-
- Beauvoit B, Rigoulet M, Guerin B, Canioni P. Polyphosphates as a source of high energy phosphates in yeast mitochondria: A 31 P NMR study. FEBS Letters. 1989;252:17–21. doi: 10.1016/0014-5793(89)80882-8. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

