SAveRUNNER: A network-based algorithm for drug repurposing and its application to COVID-19
- PMID: 33544720
- PMCID: PMC7891752
- DOI: 10.1371/journal.pcbi.1008686
SAveRUNNER: A network-based algorithm for drug repurposing and its application to COVID-19
Abstract
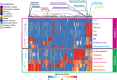
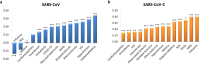
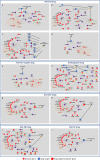
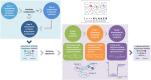
The novelty of new human coronavirus COVID-19/SARS-CoV-2 and the lack of effective drugs and vaccines gave rise to a wide variety of strategies employed to fight this worldwide pandemic. Many of these strategies rely on the repositioning of existing drugs that could shorten the time and reduce the cost compared to de novo drug discovery. In this study, we presented a new network-based algorithm for drug repositioning, called SAveRUNNER (Searching off-lAbel dRUg aNd NEtwoRk), which predicts drug-disease associations by quantifying the interplay between the drug targets and the disease-specific proteins in the human interactome via a novel network-based similarity measure that prioritizes associations between drugs and diseases locating in the same network neighborhoods. Specifically, we applied SAveRUNNER on a panel of 14 selected diseases with a consolidated knowledge about their disease-causing genes and that have been found to be related to COVID-19 for genetic similarity (i.e., SARS), comorbidity (e.g., cardiovascular diseases), or for their association to drugs tentatively repurposed to treat COVID-19 (e.g., malaria, HIV, rheumatoid arthritis). Focusing specifically on SARS subnetwork, we identified 282 repurposable drugs, including some the most rumored off-label drugs for COVID-19 treatments (e.g., chloroquine, hydroxychloroquine, tocilizumab, heparin), as well as a new combination therapy of 5 drugs (hydroxychloroquine, chloroquine, lopinavir, ritonavir, remdesivir), actually used in clinical practice. Furthermore, to maximize the efficiency of putative downstream validation experiments, we prioritized 24 potential anti-SARS-CoV repurposable drugs based on their network-based similarity values. These top-ranked drugs include ACE-inhibitors, monoclonal antibodies (e.g., anti-IFNγ, anti-TNFα, anti-IL12, anti-IL1β, anti-IL6), and thrombin inhibitors. Finally, our findings were in-silico validated by performing a gene set enrichment analysis, which confirmed that most of the network-predicted repurposable drugs may have a potential treatment effect against human coronavirus infections.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Tang X, Wu C, Li X, Song Y, Yao X, Wu X, et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev [Internet]. [cited 2020 Mar 19]; Available from: https://academic.oup.com/nsr/advance-article/doi/10.1093/nsr/nwaa036/577.... - DOI - PMC - PubMed
-
- Raoult D, Zumla A, Locatelli F, Ippolito G, Kroemer G. Coronavirus infections: Epidemiological, clinical and immunological features and hypotheses. Cell Stress [Internet]. 2020. March 2 [cited 2020 Apr 6]; Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7064018/ 10.15698/cst2020.04.216 - DOI - PMC - PubMed
-
- Coronavirus COVID-19 (2019-nCoV) [Internet]. [cited 2020 Mar 19]. Available from: https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

