TCR+/BCR+ dual-expressing cells and their associated public BCR clonotype are not enriched in type 1 diabetes
- PMID: 33545036
- PMCID: PMC8016147
- DOI: 10.1016/j.cell.2020.11.035
TCR+/BCR+ dual-expressing cells and their associated public BCR clonotype are not enriched in type 1 diabetes
Abstract
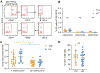
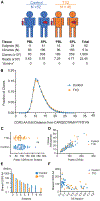
Ahmed and colleagues recently described a novel hybrid lymphocyte expressing both a B and T cell receptor, termed double expresser (DE) cells. DE cells in blood of type 1 diabetes (T1D) subjects were present at increased numbers and enriched for a public B cell clonotype. Here, we attempted to reproduce these findings. While we could identify DE cells by flow cytometry, we found no association between DE cell frequency and T1D status. We were unable to identify the reported public B cell clone, or any similar clone, in bulk B cells or sorted DE cells from T1D subjects or controls. We also did not observe increased usage of the public clone VH or DH genes in B cells or in sorted DE cells. Taken together, our findings suggest that DE cells and their alleged public clonotype are not enriched in T1D. This Matters Arising paper is in response to Ahmed et al. (2019), published in Cell. See also the response by Ahmed et al. (2021), published in this issue.
Keywords: HIRN; HPAP; Human Islet Research Network; Human Pancreas Analysis Program; T1D; immune repertoire; immunoglobulin; immunophenotyping; public clone; rare-event detection; type 1 diabetes.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





Comment in
-
A reply to "TCR+/BCR+ dual-expressing cells and their associated public BCR clonotype are not enriched in type 1 diabetes".Cell. 2021 Feb 4;184(3):840-843. doi: 10.1016/j.cell.2020.11.036. Cell. 2021. PMID: 33545037 Free PMC article.
Comment on
-
A Public BCR Present in a Unique Dual-Receptor-Expressing Lymphocyte from Type 1 Diabetes Patients Encodes a Potent T Cell Autoantigen.Cell. 2019 May 30;177(6):1583-1599.e16. doi: 10.1016/j.cell.2019.05.007. Cell. 2019. PMID: 31150624 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

