Mutant p53 suppresses innate immune signaling to promote tumorigenesis
- PMID: 33545063
- PMCID: PMC8044023
- DOI: 10.1016/j.ccell.2021.01.003
Mutant p53 suppresses innate immune signaling to promote tumorigenesis
Abstract
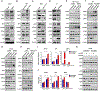
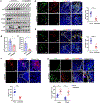
Mutant p53 (mtp53) proteins can exert cancer-promoting gain-of-function activities. We report a mechanism by which mtp53 suppresses both cell-autonomous and non-cell-autonomous signaling to promote cancer cell survival and evasion of tumor immune surveillance. Mtp53 interferes with the function of the cytoplasmic DNA sensing machinery, cGAS-STING-TBK1-IRF3, that activates the innate immune response. Mtp53, but not wild-type p53, binds to TANK-binding protein kinase 1 (TBK1) and prevents the formation of a trimeric complex between TBK1, STING, and IRF3, which is required for activation, nuclear translocation, and transcriptional activity of IRF3. Inactivation of innate immune signaling by mtp53 alters cytokine production, resulting in immune evasion. Restoring TBK1 signaling is sufficient to bypass mtp53 and lead to restored immune cell function and cancer cell eradication. This work is of translational interest because therapeutic approaches that restore TBK1 function could potentially reactivate immune surveillance and eliminate mtp53 tumors.
Keywords: IRF3; STING; TBK1; immune evasion; innate immune signaling; mutant p53.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





Comment in
-
Mutant p53 chills tumours by turning off cGAS.Nat Rev Immunol. 2021 Mar;21(3):134-135. doi: 10.1038/s41577-021-00518-x. Nat Rev Immunol. 2021. PMID: 33589807 No abstract available.
References
-
- Ablasser A, and Chen ZJ (2019). cGAS in action: Expanding roles in immunity and inflammation. Science 363. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

