Sequence-specific and multiplex detection of COVID-19 virus (SARS-CoV-2) using proofreading enzyme-mediated probe cleavage coupled with isothermal amplification
- PMID: 33545551
- PMCID: PMC7842130
- DOI: 10.1016/j.bios.2021.113041
Sequence-specific and multiplex detection of COVID-19 virus (SARS-CoV-2) using proofreading enzyme-mediated probe cleavage coupled with isothermal amplification
Abstract
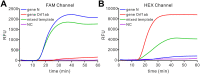
The outbreak of COVID-19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been challenging human health worldwide. Loop-mediated isothermal amplification (LAMP) has been promptly applied to the detection of SARS-CoV-2 owing to its high amplification efficacy and less requirement of the thermal cycler. However, the vast majority of these LAMP-based assays depend on the non-specific detection of LAMP products, which can not discern the undesirable amplificons, likely to yield unreliable results. Herein, a sequence-specific LAMP assay was reported to detect SARS-CoV-2 using proofreading enzyme-mediated probe cleavage (named Proofman), which could realize real-time and visual detection without uncapping. This assay, introducing a proofreading enzyme and the fluorogenic probe to reverse-transcription LAMP (RT-Proofman-LAMP), can specifically detect the SARS-CoV-2 RNA with a detection limit of 100 copies. In addition to the real-time analysis, the assay is capable of endpoint visualization under a transilluminator within 50 min, providing a convenient reporting manner under the setting of point-of-care testing (POCT). In combination with different fluorophores, the one-pot multiplex assay was successfully achieved to detect multiple targets of SARS-CoV-2 and inner control simultaneously. In summary, the development of RT-Proofman-LAMP offers a versatile and highly-specific method for fast field screening and laboratory testing of SARS-CoV-2, making it a promising platform in COVID-19 diagnosis.
Keywords: COVID-19; LAMP; Proofman; SARS-CoV-2; Sequence-specific.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection.Biosens Bioelectron. 2021 Jan 15;172:112766. doi: 10.1016/j.bios.2020.112766. Epub 2020 Oct 26. Biosens Bioelectron. 2021. PMID: 33126177 Free PMC article.
-
Multisite Clinical Validation of Isothermal Amplification-Based SARS-CoV-2 Detection Assays Using Different Sampling Strategies.Microbiol Spectr. 2021 Oct 31;9(2):e0084621. doi: 10.1128/Spectrum.00846-21. Epub 2021 Oct 20. Microbiol Spectr. 2021. PMID: 34668736 Free PMC article.
-
Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19.Biosens Bioelectron. 2020 Oct 15;166:112437. doi: 10.1016/j.bios.2020.112437. Epub 2020 Jul 15. Biosens Bioelectron. 2020. PMID: 32692666 Free PMC article.
-
Loop-mediated isothermal amplification (LAMP): An effective molecular point-of-care technique for the rapid diagnosis of coronavirus SARS-CoV-2.Rev Med Virol. 2021 Nov;31(6):e2215. doi: 10.1002/rmv.2215. Epub 2021 Jan 21. Rev Med Virol. 2021. PMID: 33476080 Free PMC article. Review.
-
Nucleic Acid Testing of SARS-CoV-2.Int J Mol Sci. 2021 Jun 7;22(11):6150. doi: 10.3390/ijms22116150. Int J Mol Sci. 2021. PMID: 34200331 Free PMC article. Review.
Cited by
-
Detection of Soybean-Derived Components in Dairy Products Using Proofreading Enzyme-Mediated Probe Cleavage Coupled with Ladder-Shape Melting Temperature Isothermal Amplification (Proofman-LMTIA).Molecules. 2023 Feb 10;28(4):1685. doi: 10.3390/molecules28041685. Molecules. 2023. PMID: 36838673 Free PMC article.
-
Batteryless wireless magnetostrictive Fe30Co70/Ni clad plate for human coronavirus 229E detection.Sens Actuators A Phys. 2023 Jan 1;349:114052. doi: 10.1016/j.sna.2022.114052. Epub 2022 Nov 24. Sens Actuators A Phys. 2023. PMID: 36447950 Free PMC article.
-
Isothermal amplification-assisted diagnostics for COVID-19.Biosens Bioelectron. 2022 Jun 1;205:114101. doi: 10.1016/j.bios.2022.114101. Epub 2022 Feb 17. Biosens Bioelectron. 2022. PMID: 35202984 Free PMC article. Review.
-
Monolithic, 3D-printed lab-on-disc platform for multiplexed molecular detection of SARS-CoV-2.Sens Actuators B Chem. 2022 Jan 15;351:130998. doi: 10.1016/j.snb.2021.130998. Epub 2021 Oct 27. Sens Actuators B Chem. 2022. PMID: 34725537 Free PMC article.
-
Multiplexed lateral flow assay integrated with orthogonal CRISPR-Cas system for SARS-CoV-2 detection.Sens Actuators B Chem. 2022 Nov 15;371:132537. doi: 10.1016/j.snb.2022.132537. Epub 2022 Aug 23. Sens Actuators B Chem. 2022. PMID: 36032355 Free PMC article.
References
-
- Chen G., Chen R., Ding S., Li M., Wang J., Zou J., Du F., Dong J., Cui X., Huang X., Deng Y., Tang Z. Recombinase assisted loop-mediated isothermal DNA amplification. Analyst. 2020;145(2):440–444. - PubMed
-
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) - PMC - PubMed
-
- Jung Y., Park G.S., Moon J.H., Ku K., Beak S.H., Lee C.S., Kim S., Park E.C., Park D., Lee J.H., Byeon C.W., Lee J.J., Maeng J.S., Kim S.J., Kim S.I., Kim B.T., Lee M.J., Kim H.G. Comparative analysis of primer-probe sets for RT-qPCR of COVID-19 causative virus (SARS-CoV-2) ACS Infect. Dis. 2020;6(9):2513–2523. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

