The First Whole Genome Sequence and Characterisation of Avian Nephritis Virus Genotype 3
- PMID: 33546203
- PMCID: PMC7913312
- DOI: 10.3390/v13020235
The First Whole Genome Sequence and Characterisation of Avian Nephritis Virus Genotype 3
Abstract
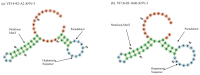
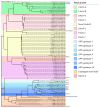
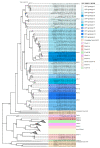
Avian nephritis virus (ANV) is classified in the Avastroviridae family with disease associations with nephritis, uneven flock growth and runting stunting syndrome (RSS) in chicken and turkey flocks, and other avian species. The whole genome of ANV genotype 3 (ANV-3) of 6959 nucleotides including the untranslated 5' and 3' regions and polyadenylated tail was detected in a metagenomic virome investigation of RSS-affected chicken broiler flocks. This report characterises the ANV-3 genome, identifying partially overlapping open reading frames (ORFs), ORF1a and ORF1b, and an opposing secondary pseudoknot prior to a ribosomal frameshift stemloop structure, with a separate ORF2, whilst observing conserved astrovirus motifs. Phylogenetic analysis of the Avastroviridae whole genome and ORF2 capsid polyprotein classified the first complete whole genome of ANV-3 within Avastroviridae genogroup 2.
Keywords: astrovirus; avian nephritis virus; genotyping and recombination; molecular characterisation; phylogenetics.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Asplin F.D. Duck Hepatitis. Vet. Rec. 1965;77:487–488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

