Long-Range PCR-Based NGS Applications to Diagnose Mendelian Retinal Diseases
- PMID: 33546218
- PMCID: PMC7913364
- DOI: 10.3390/ijms22041508
Long-Range PCR-Based NGS Applications to Diagnose Mendelian Retinal Diseases
Abstract
The purpose of this study was to develop a flexible, cost-efficient, next-generation sequencing (NGS) protocol for genetic testing. Long-range polymerase chain reaction (PCR) amplicons of up to 20 kb in size were designed to amplify entire genomic regions for a panel (n = 35) of inherited retinal disease (IRD)-associated loci. Amplicons were pooled and sequenced by NGS. The analysis was applied to 227 probands diagnosed with IRD: (A) 108 previously molecularly diagnosed, (B) 94 without previous genetic testing, and (C) 25 undiagnosed after whole-exome sequencing (WES). The method was validated with 100% sensitivity on cohort A. Long-range PCR-based sequencing revealed likely causative variant(s) in 51% and 24% of proband from cohorts B and C, respectively. Breakpoints of 3 copy number variants (CNVs) could be characterized. Long-range PCR libraries spike-in extended coverage of WES. Read phasing confirmed compound heterozygosity in 5 probands. The proposed sequencing protocol provided deep coverage of the entire gene, including intronic and promoter regions. Our method can be used (i) as a first-tier assay to reduce genetic testing costs, (ii) to elucidate missing heritability cases, (iii) to characterize breakpoints of CNVs at nucleotide resolution, (iv) to extend WES data to non-coding regions by spiking-in long-range PCR libraries, and (v) to help with phasing of candidate variants.
Keywords: ABCA4; BEST1; CNV; NGS; PRPH2; diagnostics; genetic testing; long-range PCR; missing heritability; phasing; retinal diseases; sequencing.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

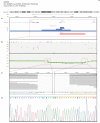



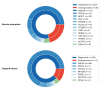
References
-
- Tiwari A., Lemke J., Altmueller J., Thiele H., Glaus E., Fleischhauer J., Nürnberg P., Neidhardt J., Berger W. Identification of Novel and Recurrent Disease-Causing Mutations in Retinal Dystrophies Using Whole Exome Sequencing (WES): Benefits and Limitations. PLoS ONE. 2016;11:e0158692. doi: 10.1371/journal.pone.0158692. - DOI - PMC - PubMed
-
- Toulis V., Cortés-González V., De Castro-Miró M., Sallum J.M.F., Català-Mora J., Villanueva-Mendoza C., Ciccioli M., Gonzàlez-Duarte R., Valero R., Marfany G. Increasing the Genetic Diagnosis Yield in Inherited Retinal Dystrophies: Assigning Pathogenicity to Novel Non-canonical Splice Site Variants. Genes. 2020;11:378. doi: 10.3390/genes11040378. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

