Mucosal-Associated Invariant T (MAIT) Cells Are Highly Activated and Functionally Impaired in COVID-19 Patients
- PMID: 33546489
- PMCID: PMC7913667
- DOI: 10.3390/v13020241
Mucosal-Associated Invariant T (MAIT) Cells Are Highly Activated and Functionally Impaired in COVID-19 Patients
Abstract
Coronavirus disease 2019 (COVID-19), caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), comprises mild courses of disease as well as progression to severe disease, characterised by lung and other organ failure. The immune system is considered to play a crucial role for the pathogenesis of COVID-19, although especially the contribution of innate-like T cells remains poorly understood. Here, we analysed the phenotype and function of mucosal-associated invariant T (MAIT) cells, innate-like T cells with potent antimicrobial effector function, in patients with mild and severe COVID-19 by multicolour flow cytometry. Our data indicate that MAIT cells are highly activated in patients with COVID-19, irrespective of the course of disease, and express high levels of proinflammatory cytokines such as IL-17A and TNFα ex vivo. Of note, expression of the activation marker HLA-DR positively correlated with SAPS II score, a measure of disease severity. Upon MAIT cell-specific in vitro stimulation, MAIT cells however failed to upregulate expression of the cytokines IL-17A and TNFα, as well as cytolytic proteins, that is, granzyme B and perforin. Thus, our data point towards an altered cytokine expression profile alongside an impaired antibacterial and antiviral function of MAIT cells in COVID-19 and thereby contribute to the understanding of COVID-19 immunopathogenesis.
Keywords: COVID-19; SARS-CoV-2; mucosal-associated invariant T (MAIT) cells.
Conflict of interest statement
The authors declare no conflict of interest with regards to the present manuscript.
Figures
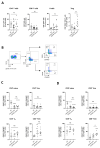
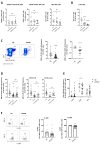
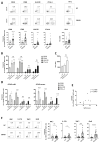

References
-
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. - DOI - PMC - PubMed
-
- The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:145–151. - PubMed
-
- Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., Cuomo-Dannenburg G., Thompson H., Walker P.G.T., Fu H., et al. Estimates of the severity of coronavirus disease 2019: A model-based analysis. Lancet Infect. Dis. 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

