Targeted enrichment of novel chloroplast-based probes reveals a large-scale phylogeny of 412 bamboos
- PMID: 33546593
- PMCID: PMC7863319
- DOI: 10.1186/s12870-020-02779-5
Targeted enrichment of novel chloroplast-based probes reveals a large-scale phylogeny of 412 bamboos
Abstract
Background: The subfamily Bambusoideae belongs to the grass family Poaceae and has significant roles in culture, economy, and ecology. However, the phylogenetic relationships based on large-scale chloroplast genomes (CpGenomes) were elusive. Moreover, most of the chloroplast DNA sequencing methods cannot meet the requirements of large-scale CpGenome sequencing, which greatly limits and impedes the in-depth research of plant genetics and evolution.
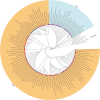
Results: To develop a set of bamboo probes, we used 99 high-quality CpGenomes with 6 bamboo CpGenomes as representative species for the probe design, and assembled 15 M unique sequences as the final pan-chloroplast genome. A total of 180,519 probes for chloroplast DNA fragments were designed and synthesized by a novel hybridization-based targeted enrichment approach. Another 468 CpGenomes were selected as test data to verify the quality of the newly synthesized probes and the efficiency of the probes for chloroplast capture. We then successfully applied the probes to synthesize, enrich, and assemble 358 non-redundant CpGenomes of woody bamboo in China. Evaluation analysis showed the probes may be applicable to chloroplasts in Magnoliales, Pinales, Poales et al. Moreover, we reconstructed a phylogenetic tree of 412 bamboos (358 in-house and 54 published), supporting a non-monophyletic lineage of the genus Phyllostachys. Additionally, we shared our data by uploading a dataset of bamboo CpGenome into CNGB ( https://db.cngb.org/search/project/CNP0000502/ ) to enrich resources and promote the development of bamboo phylogenetics.
Conclusions: The development of the CpGenome enrichment pipeline and its performance on bamboos recommended an inexpensive, high-throughput, time-saving and efficient CpGenome sequencing strategy, which can be applied to facilitate the phylogenetics analysis of most green plants.
Keywords: Bamboo phylogeny; Bambusoideae; Chloroplast; Probe; Targeted enrichment.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Evolution of the bamboos (Bambusoideae; Poaceae): a full plastome phylogenomic analysis.BMC Evol Biol. 2015 Mar 18;15:50. doi: 10.1186/s12862-015-0321-5. BMC Evol Biol. 2015. PMID: 25887467 Free PMC article.
-
High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae).PLoS One. 2011;6(5):e20596. doi: 10.1371/journal.pone.0020596. Epub 2011 May 31. PLoS One. 2011. PMID: 21655229 Free PMC article.
-
Higher level phylogenetic relationships within the bamboos (Poaceae: Bambusoideae) based on five plastid markers.Mol Phylogenet Evol. 2013 May;67(2):404-13. doi: 10.1016/j.ympev.2013.02.005. Epub 2013 Feb 20. Mol Phylogenet Evol. 2013. PMID: 23454093
-
Limitations, progress and prospects of application of biotechnological tools in improvement of bamboo-a plant with extraordinary qualities.Physiol Mol Biol Plants. 2013 Jan;19(1):21-41. doi: 10.1007/s12298-012-0147-1. Physiol Mol Biol Plants. 2013. PMID: 24381435 Free PMC article. Review.
-
Poaceae Chloroplast Genome Sequencing: Great Leap Forward in Recent Ten Years.Curr Genomics. 2023 Feb 14;23(6):369-384. doi: 10.2174/1389202924666221201140603. Curr Genomics. 2023. PMID: 37920556 Free PMC article. Review.
Cited by
-
The complete chloroplast genome of Artemisia selengensis Turcz var. shansiensis (Qinghao in Chinese).Mitochondrial DNA B Resour. 2025 Jun 18;10(7):620-625. doi: 10.1080/23802359.2025.2519250. eCollection 2025. Mitochondrial DNA B Resour. 2025. PMID: 40547542 Free PMC article.
-
Comparative analyses of chloroplast genomes in 'Red Fuji' apples: low rate of chloroplast genome mutations.PeerJ. 2022 Feb 21;10:e12927. doi: 10.7717/peerj.12927. eCollection 2022. PeerJ. 2022. PMID: 35223207 Free PMC article.
-
Multi-omics signatures of diverse plant callus cultures.Plant Biotechnol (Tokyo). 2024 Sep 25;41(3):309-314. doi: 10.5511/plantbiotechnology.24.0719a. Plant Biotechnol (Tokyo). 2024. PMID: 40115769 Free PMC article.
References
-
- Soreng RJ, et al. A worldwide phylogenetic classification of the Poaceae (Gramineae) II: an update and a comparison of two 2015 classifications. J Syst Evol. 2017;55:259–290. doi: 10.1111/jse.12262. - DOI
-
- Stapleton C, Chonghaile GN, Hodkinson TR. Molecular phylogeny of Asian woody bamboos: Review for the Flora of China. Bamboo Sci Culture. 2009;22:5–25. https://bamboo.org/ABSJournalArchive/ABSJournal-vol22/BSC%2022%20Staplet....
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

