Diel transcriptional oscillations of light-sensitive regulatory elements in open-ocean eukaryotic plankton communities
- PMID: 33547239
- PMCID: PMC8017926
- DOI: 10.1073/pnas.2011038118
Diel transcriptional oscillations of light-sensitive regulatory elements in open-ocean eukaryotic plankton communities
Abstract
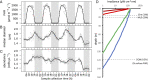
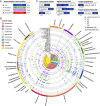
The 24-h cycle of light and darkness governs daily rhythms of complex behaviors across all domains of life. Intracellular photoreceptors sense specific wavelengths of light that can reset the internal circadian clock and/or elicit distinct phenotypic responses. In the surface ocean, microbial communities additionally modulate nonrhythmic changes in light quality and quantity as they are mixed to different depths. Here, we show that eukaryotic plankton in the North Pacific Subtropical Gyre transcribe genes encoding light-sensitive proteins that may serve as light-activated transcription factors, elicit light-driven electrical/chemical cascades, or initiate secondary messenger-signaling cascades. Overall, the protistan community relies on blue light-sensitive photoreceptors of the cryptochrome/photolyase family, and proteins containing the Light-Oxygen-Voltage (LOV) domain. The greatest diversification occurred within Haptophyta and photosynthetic stramenopiles where the LOV domain was combined with different DNA-binding domains and secondary signal-transduction motifs. Flagellated protists utilize green-light sensory rhodopsins and blue-light helmchromes, potentially underlying phototactic/photophobic and other behaviors toward specific wavelengths of light. Photoreceptors such as phytochromes appear to play minor roles in the North Pacific Subtropical Gyre. Transcript abundance of environmental light-sensitive protein-encoding genes that display diel patterns are found to primarily peak at dawn. The exceptions are the LOV-domain transcription factors with peaks in transcript abundances at different times and putative phototaxis photoreceptors transcribed throughout the day. Together, these data illustrate the diversity of light-sensitive proteins that may allow disparate groups of protists to respond to light and potentially synchronize patterns of growth, division, and mortality within the dynamic ocean environment.
Keywords: diel cycles; metatranscriptomics; microbial eukaryotes; oligotrophic gyre; photoreceptors.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Roenneberg T., Merrow M., Circadian clocks - The fall and rise of physiology. Nat. Rev. Mol. Cell Biol. 6, 965–971 (2005). - PubMed
-
- Wijnen H., Young M. W., Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 40, 409–448 (2006). - PubMed
-
- Noordally Z. B., Millar A. J., Clocks in algae. Biochemistry 54, 171–183 (2015). - PubMed
-
- Somers D. E., Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282, 1488–1490 (1998). - PubMed
-
- Cashmore A. R., Cryptochromes: Enabling plants and animals to determine circadian time. Cell 114, 537–543 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

