Sensing soluble uric acid by Naip1-Nlrp3 platform
- PMID: 33547278
- PMCID: PMC7864962
- DOI: 10.1038/s41419-021-03445-w
Sensing soluble uric acid by Naip1-Nlrp3 platform
Abstract
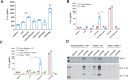
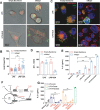
Uric acid (UA), a product of purine nucleotide degradation able to initiate an immune response, represents a breakpoint in the evolutionary history of humans, when uricase, the enzyme required for UA cleavage, was lost. Despite being inert in human cells, UA in its soluble form (sUA) can increase the level of interleukin-1β (IL-1β) in murine macrophages. We, therefore, hypothesized that the recognition of sUA is achieved by the Naip1-Nlrp3 inflammasome platform. Through structural modelling predictions and transcriptome and functional analyses, we found that murine Naip1 expression in human macrophages induces IL-1β expression, fatty acid production and an inflammation-related response upon sUA stimulation, a process reversed by the pharmacological and genetic inhibition of Nlrp3. Moreover, molecular interaction experiments showed that Naip1 directly recognizes sUA. Accordingly, Naip may be the sUA receptor lost through the human evolutionary process, and a better understanding of its recognition may lead to novel anti-hyperuricaemia therapies.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

