Galectin-9 interacts with PD-1 and TIM-3 to regulate T cell death and is a target for cancer immunotherapy
- PMID: 33547304
- PMCID: PMC7864927
- DOI: 10.1038/s41467-021-21099-2
Galectin-9 interacts with PD-1 and TIM-3 to regulate T cell death and is a target for cancer immunotherapy
Abstract
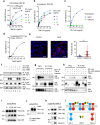
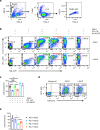
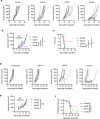
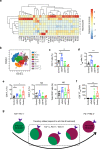
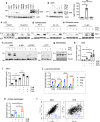
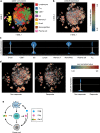
The two T cell inhibitory receptors PD-1 and TIM-3 are co-expressed during exhausted T cell differentiation, and recent evidence suggests that their crosstalk regulates T cell exhaustion and immunotherapy efficacy; however, the molecular mechanism is unclear. Here we show that PD-1 contributes to the persistence of PD-1+TIM-3+ T cells by binding to the TIM-3 ligand galectin-9 (Gal-9) and attenuates Gal-9/TIM-3-induced cell death. Anti-Gal-9 therapy selectively expands intratumoral TIM-3+ cytotoxic CD8 T cells and immunosuppressive regulatory T cells (Treg cells). The combination of anti-Gal-9 and an agonistic antibody to the co-stimulatory receptor GITR (glucocorticoid-induced tumor necrosis factor receptor-related protein) that depletes Treg cells induces synergistic antitumor activity. Gal-9 expression and secretion are promoted by interferon β and γ, and high Gal-9 expression correlates with poor prognosis in multiple human cancers. Our work uncovers a function for PD-1 in exhausted T cell survival and suggests Gal-9 as a promising target for immunotherapy.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

