Lung microbiota associations with clinical features of COPD in the SPIROMICS cohort
- PMID: 33547327
- PMCID: PMC7865064
- DOI: 10.1038/s41522-021-00185-9
Lung microbiota associations with clinical features of COPD in the SPIROMICS cohort
Abstract
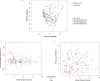
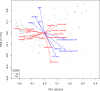
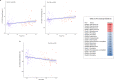
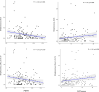
Chronic obstructive pulmonary disease (COPD) is heterogeneous in development, progression, and phenotypes. Little is known about the lung microbiome, sampled by bronchoscopy, in milder COPD and its relationships to clinical features that reflect disease heterogeneity (lung function, symptom burden, and functional impairment). Using bronchoalveolar lavage fluid collected from 181 never-smokers and ever-smokers with or without COPD (GOLD 0-2) enrolled in the SubPopulations and InteRmediate Outcome Measures In COPD Study (SPIROMICS), we find that lung bacterial composition associates with several clinical features, in particular bronchodilator responsiveness, peak expiratory flow rate, and forced expiratory flow rate between 25 and 75% of FVC (FEF25-75). Measures of symptom burden (COPD Assessment Test) and functional impairment (six-minute walk distance) also associate with disparate lung microbiota composition. Drivers of these relationships include members of the Streptococcus, Prevotella, Veillonella, Staphylococcus, and Pseudomonas genera. Thus, lung microbiota differences may contribute to airway dysfunction and airway disease in milder COPD.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Lung Microbiota and Metabolites Collectively Associate with Clinical Outcomes in Milder Stage Chronic Obstructive Pulmonary Disease.Am J Respir Crit Care Med. 2022 Aug 15;206(4):427-439. doi: 10.1164/rccm.202110-2241OC. Am J Respir Crit Care Med. 2022. PMID: 35536732 Free PMC article.
-
Analysis of the lung microbiome in the "healthy" smoker and in COPD.PLoS One. 2011 Feb 22;6(2):e16384. doi: 10.1371/journal.pone.0016384. PLoS One. 2011. PMID: 21364979 Free PMC article. Clinical Trial.
-
Analysis of the bronchoalveolar lavage fluid microbial flora in COPD patients at different lung function during acute exacerbation.Sci Rep. 2025 Apr 16;15(1):13179. doi: 10.1038/s41598-025-96746-5. Sci Rep. 2025. PMID: 40240456 Free PMC article.
-
COPD and the microbiome.Respirology. 2016 May;21(4):590-9. doi: 10.1111/resp.12732. Epub 2016 Jan 27. Respirology. 2016. PMID: 26852737 Review.
-
The lung microbiome dynamics between stability and exacerbation in chronic obstructive pulmonary disease (COPD): Current perspectives.Respir Med. 2019 Oct;157:1-6. doi: 10.1016/j.rmed.2019.08.012. Epub 2019 Aug 21. Respir Med. 2019. PMID: 31450162 Review.
Cited by
-
Nutritional immunity: the impact of metals on lung immune cells and the airway microbiome during chronic respiratory disease.Respir Res. 2021 Apr 29;22(1):133. doi: 10.1186/s12931-021-01722-y. Respir Res. 2021. PMID: 33926483 Free PMC article. Review.
-
Comparative analysis of the bronchoalveolar microbiome in Portuguese patients with different chronic lung disorders.Sci Rep. 2021 Jul 22;11(1):15042. doi: 10.1038/s41598-021-94468-y. Sci Rep. 2021. PMID: 34294826 Free PMC article.
-
Informatic analysis of the pulmonary microecology in non-cystic fibrosis bronchiectasis at three different stages.Open Life Sci. 2022 Feb 28;17(1):107-120. doi: 10.1515/biol-2022-0014. eCollection 2022. Open Life Sci. 2022. PMID: 35291562 Free PMC article.
-
Lung Microbiota and Metabolites Collectively Associate with Clinical Outcomes in Milder Stage Chronic Obstructive Pulmonary Disease.Am J Respir Crit Care Med. 2022 Aug 15;206(4):427-439. doi: 10.1164/rccm.202110-2241OC. Am J Respir Crit Care Med. 2022. PMID: 35536732 Free PMC article.
-
ERS International Congress, Madrid, 2019: highlights from the Airway Diseases, Asthma and COPD Assembly.ERJ Open Res. 2020 Feb 17;6(1):00341-2019. doi: 10.1183/23120541.00341-2019. eCollection 2020 Jan. ERJ Open Res. 2020. PMID: 32083111 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- HHSN268200900019C/HL/NHLBI NIH HHS/United States
- U24 HL141762/HL/NHLBI NIH HHS/United States
- P30 ES005605/ES/NIEHS NIH HHS/United States
- R01 HL121774/HL/NHLBI NIH HHS/United States
- HHSN268200900015C/HL/NHLBI NIH HHS/United States
- HHSN268200900016C/HL/NHLBI NIH HHS/United States
- U01 HL137880/HL/NHLBI NIH HHS/United States
- HHSN268200900013C/HL/NHLBI NIH HHS/United States
- R01 AI129958/AI/NIAID NIH HHS/United States
- HHSN268200900014C/HL/NHLBI NIH HHS/United States
- K24 HL138188/HL/NHLBI NIH HHS/United States
- R03 HL138310/HL/NHLBI NIH HHS/United States
- K23 HL123778/HL/NHLBI NIH HHS/United States
- K24 HL137013/HL/NHLBI NIH HHS/United States
- HHSN268200900018C/HL/NHLBI NIH HHS/United States
- P30 DK054759/DK/NIDDK NIH HHS/United States
- HHSN268200900017C/HL/NHLBI NIH HHS/United States
- HHSN268200900020C/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

