Multimodal deep learning models for early detection of Alzheimer's disease stage
- PMID: 33547343
- PMCID: PMC7864942
- DOI: 10.1038/s41598-020-74399-w
Multimodal deep learning models for early detection of Alzheimer's disease stage
Abstract
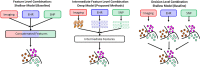
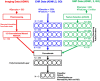
Most current Alzheimer's disease (AD) and mild cognitive disorders (MCI) studies use single data modality to make predictions such as AD stages. The fusion of multiple data modalities can provide a holistic view of AD staging analysis. Thus, we use deep learning (DL) to integrally analyze imaging (magnetic resonance imaging (MRI)), genetic (single nucleotide polymorphisms (SNPs)), and clinical test data to classify patients into AD, MCI, and controls (CN). We use stacked denoising auto-encoders to extract features from clinical and genetic data, and use 3D-convolutional neural networks (CNNs) for imaging data. We also develop a novel data interpretation method to identify top-performing features learned by the deep-models with clustering and perturbation analysis. Using Alzheimer's disease neuroimaging initiative (ADNI) dataset, we demonstrate that deep models outperform shallow models, including support vector machines, decision trees, random forests, and k-nearest neighbors. In addition, we demonstrate that integrating multi-modality data outperforms single modality models in terms of accuracy, precision, recall, and meanF1 scores. Our models have identified hippocampus, amygdala brain areas, and the Rey Auditory Verbal Learning Test (RAVLT) as top distinguished features, which are consistent with the known AD literature.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

