Efficient Gene Stacking in Rice Using the GAANTRY System
- PMID: 33547973
- PMCID: PMC7867672
- DOI: 10.1186/s12284-021-00460-5
Efficient Gene Stacking in Rice Using the GAANTRY System
Abstract
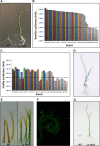
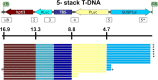
Genetic engineering of rice provides a means for improving rice grain quality and yield, and the introduction and expression of multiple genes can produce new traits that would otherwise be difficult to obtain through conventional breeding. GAANTRY (Gene Assembly in Agrobacterium by Nucleic acid Transfer using Recombinase technologY) was previously shown to be a precise and robust system to stably stack ten genes (28 kilobases (kb)) within an Agrobacterium virulence plasmid Transfer-DNA (T-DNA) and obtain high-quality Arabidopsis and potato transgenic events. To determine whether the GAANTRY system can be used to engineer a monocotyledonous crop, two new T-DNA constructs, carrying five (16.9 kb) or eleven (37.4 kb) cargo sequences were assembled and transformed into rice. Characterization of 53 independent transgenic events demonstrated that more than 50% of the plants carried all of the desired cargo sequences and exhibited the introduced traits. Additionally, more than 18% of the lines were high-quality events containing a single copy of the introduced transgenes and were free of sequences from outside of the T-DNA. Therefore, GAANTRY provides a simple, precise and versatile tool for transgene stacking in rice and potentially other cereal grain crops.
Keywords: Agrobacterium; Gene stacking; Genetic engineering; Oryza sativa; Site-specific recombinase.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Bao J. Rice: chemistry and technology. 2018. Biotechnology for rice grain quality improvement.
-
- Biswal AK, Shamim M, Cruzado K, et al. Rice production worldwide. 2017. Role of biotechnology in rice production.
-
- Brar DS, Khush GS. Agricultural sustainability. 2013. Biotechnological approaches for increasing productivity and sustainability of rice production.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

