Lnc-ORA interacts with microRNA-532-3p and IGF2BP2 to inhibit skeletal muscle myogenesis
- PMID: 33548229
- PMCID: PMC8289116
- DOI: 10.1016/j.jbc.2021.100376
Lnc-ORA interacts with microRNA-532-3p and IGF2BP2 to inhibit skeletal muscle myogenesis
Abstract
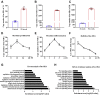
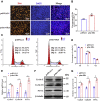
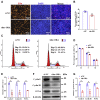
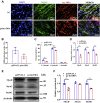
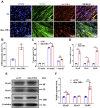
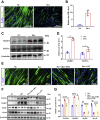
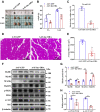
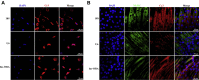
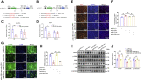
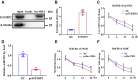
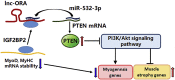
Skeletal muscle is one of the most important organs of the animal body. Long noncoding RNAs play a crucial role in the regulation of skeletal muscle development via several mechanisms. We recently identified obesity-related lncRNA (lnc-ORA) in a search for long noncoding RNAs that influence adipogenesis, finding it impacted adipocyte differentiation by regulating the PI3K/protein kinase B/mammalian target of rapamycin pathway. However, whether lnc-ORA has additional roles, specifically in skeletal muscle myogenesis, is not known. Here, we found that lnc-ORA was significantly differentially expressed with age in mouse skeletal muscle tissue and predominantly located in the cytoplasm. Overexpression of lnc-ORA promoted C2C12 myoblast proliferation and inhibited myoblast differentiation. In contrast, lnc-ORA knockdown repressed myoblast proliferation and facilitated myoblast differentiation. Interestingly, silencing of lnc-ORA rescued dexamethasone-induced muscle atrophy in vitro. Furthermore, adeno-associated virus 9-mediated overexpression of lnc-ORA decreased muscle mass and the cross-sectional area of muscle fiber by upregulating the levels of muscle atrophy-related genes and downregulating the levels of myogenic differentiation-related genes in vivo. Mechanistically, lnc-ORA inhibited skeletal muscle myogenesis by acting as a sponge of miR-532-3p, which targets the phosphatase and tensin homolog gene; the resultant changes in phosphatase and tensin homolog suppressed the PI3K/protein kinase B signaling pathway. In addition, lnc-ORA interacted with insulin-like growth factor 2 mRNA-binding protein 2 and reduced the stability of myogenesis genes, such as myogenic differentiation 1 and myosin heavy chain. Collectively, these findings indicate that lnc-ORA could be a novel underlying regulator of skeletal muscle development.
Keywords: PTEN/PI3K/AKT signaling pathway; insulin-like growth factor 2 mRNA-binding protein 2; lnc-ORA; miR-532-3p; myogenesis; skeletal muscle.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures
Similar articles
-
Lnc-mg is a long non-coding RNA that promotes myogenesis.Nat Commun. 2017 Mar 10;8:14718. doi: 10.1038/ncomms14718. Nat Commun. 2017. PMID: 28281528 Free PMC article.
-
LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway.J Cachexia Sarcopenia Muscle. 2019 Apr;10(2):391-410. doi: 10.1002/jcsm.12374. Epub 2019 Jan 30. J Cachexia Sarcopenia Muscle. 2019. PMID: 30701698 Free PMC article.
-
MicroRNA-351-5p mediates skeletal myogenesis by directly targeting lactamase-β and is regulated by lnc-mg.FASEB J. 2019 Feb;33(2):1911-1926. doi: 10.1096/fj.201701394RRR. Epub 2018 Sep 14. FASEB J. 2019. PMID: 30216112
-
MicroRNA in myogenesis and muscle atrophy.Curr Opin Clin Nutr Metab Care. 2013 May;16(3):258-66. doi: 10.1097/MCO.0b013e32835f81b9. Curr Opin Clin Nutr Metab Care. 2013. PMID: 23449000 Free PMC article. Review.
-
Circular RNAs and host genes act synergistically in regulating cellular processes and functions in skeletal myogenesis.Gene. 2025 Mar 10;940:149189. doi: 10.1016/j.gene.2024.149189. Epub 2024 Dec 24. Gene. 2025. PMID: 39724991 Review.
Cited by
-
The Functional Role of Long Non-Coding RNA in Myogenesis and Skeletal Muscle Atrophy.Cells. 2022 Jul 25;11(15):2291. doi: 10.3390/cells11152291. Cells. 2022. PMID: 35892588 Free PMC article. Review.
-
miRNAs Influence m6A RNA Methylation through FTO and IGF2BP2 in Pressure Overload-Induced Heart Failure.Comb Chem High Throughput Screen. 2025;28(6):1072-1084. doi: 10.2174/0113862073316437240426071022. Comb Chem High Throughput Screen. 2025. PMID: 38693740
-
lncRNA HCG11 Promotes Colorectal Cancer Cell Malignant Behaviors via Sponging miR-26b-5p.J Immunol Res. 2023 Feb 23;2023:9011232. doi: 10.1155/2023/9011232. eCollection 2023. J Immunol Res. 2023. PMID: 36874625 Free PMC article.
-
Removal of MuRF1 Increases Muscle Mass in Nemaline Myopathy Models, but Does Not Provide Functional Benefits.Int J Mol Sci. 2022 Jul 23;23(15):8113. doi: 10.3390/ijms23158113. Int J Mol Sci. 2022. PMID: 35897687 Free PMC article.
-
The role of non-coding RNAs in muscle aging: regulatory mechanisms and therapeutic potential.Front Mol Biosci. 2024 Jan 9;10:1308274. doi: 10.3389/fmolb.2023.1308274. eCollection 2023. Front Mol Biosci. 2024. PMID: 38264571 Free PMC article. Review.
References
-
- Braun T., Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell. Biol. 2011;12:349–361. - PubMed
-
- Buckingham M., Rigby P.W. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell. 2014;28:225–238. - PubMed
-
- Schiaffino S., Dyar K.A., Ciciliot S., Blaauw B., Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013;280:4294–4314. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

